 |
PDBsum entry 1f3t

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of trypanosoma brucei ornithine decarboxylase (odc) complexed with putrescine, odc's reaction product.
|
|
Structure:
|
 |
Ornithine decarboxylase. Chain: a, b, c, d. Synonym: odc. Engineered: yes
|
|
Source:
|
 |
Trypanosoma brucei. Organism_taxid: 5691. Expressed in: escherichia coli. Expression_system_taxid: 562. Other_details: modified pet-11d plasmid with his6-tag, tev protease site and t7 promotor. His6-tag was cleaved.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.237
|
R-free:
|
0.280
|
|
|
Authors:
|
 |
L.K.Jackson,H.B.Brooks,A.L.Osterman,E.J.Goldsmith,M.A.Phillips
|
Key ref:

|
 |
L.K.Jackson
et al.
(2000).
Altering the reaction specificity of eukaryotic ornithine decarboxylase.
Biochemistry,
39,
11247-11257.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
06-Jun-00
|
Release date:
|
22-Nov-00
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P07805
(DCOR_TRYBB) -
Ornithine decarboxylase from Trypanosoma brucei brucei
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
423 a.a.
378 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.17
- ornithine decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Spermine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-ornithine + H+ = putrescine + CO2
|
 |
 |
 |
 |
 |
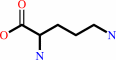 L-ornithine
L-ornithine
|
+
|
H(+)
|
=
|
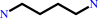 putrescine
putrescine
|
+
|
CO2
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
39:11247-11257
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Altering the reaction specificity of eukaryotic ornithine decarboxylase.
|
|
L.K.Jackson,
H.B.Brooks,
A.L.Osterman,
E.J.Goldsmith,
M.A.Phillips.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ornithine decarboxylase (ODC) catalyzes the first committed step in the
biosynthesis of polyamines, and it has been identified as a drug target for the
treatment of African sleeping sickness, caused by Trypanosoma brucei. ODC is a
pyridoxal 5'-phosphate (PLP) dependent enzyme and an obligate homodimer. X-ray
structural analysis of the complex of the T. brucei wild-type enzyme with the
product putrescine reveals two structural changes that occur upon ligand
binding: Lys-69 is displaced by putrescine and forms new interactions with
Glu-94 and Asp-88, and the side chain of Cys-360 rotates into the active site to
within 3.4 A of the imine bond. Mutation of Cys-360 to Ala or Ser reduces the
k(cat) of the decarboxylation reaction by 50- and 1000-fold, respectively.
However, HPLC analysis of the products demonstrates that the mutant enzymes
almost exclusively catalyze a decarboxylation-dependent transamination reaction
to form pyridoxamine 5-phosphate (PMP) and gamma-aminobutyraldehyde, instead of
PLP and putrescine. This side reaction arises when the decarboxylated substrate
intermediate is protonated at C4' of PLP instead of at the C(alpha) of
substrate. For the reaction catalyzed by the wild-type enzyme, this side
reaction occurs infrequently (<0.01% of the turnovers). Single turnover
analysis and multiwavelength stopped-flow spectroscopic studies suggest that for
the mutant ODCs protonation at C4' occurs either very rapidly or in a concerted
reaction with decarboxylation and that the rate-limiting step in the
steady-state reaction is Schiff base hydrolysis/product release. These studies
demonstrate a role for Cys-360 in the control of the C(alpha) protonation step
that catalyzes the formation of the physiological product putrescine. The
results further provide insight into the mechanism by which this class of
PLP-dependent enzymes controls reaction specificity.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.P.Ivanov,
A.E.Firth,
and
J.F.Atkins
(2010).
Recurrent emergence of catalytically inactive ornithine decarboxylase homologous forms that likely have regulatory function.
|
| |
J Mol Evol,
70,
289-302.
|
 |
|
|
|
|
 |
S.Albeck,
O.Dym,
T.Unger,
Z.Snapir,
Z.Bercovich,
and
C.Kahana
(2008).
Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function.
|
| |
Protein Sci,
17,
793-802.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Hu,
D.Wu,
J.Chen,
J.Ding,
H.Jiang,
and
X.Shen
(2008).
The catalytic intermediate stabilized by a "down" active site loop for diaminopimelate decarboxylase from Helicobacter pylori. Enzymatic characterization with crystal structure analysis.
|
| |
J Biol Chem,
283,
21284-21293.
|
 |
|
|
|
|
 |
R.Shah,
R.Akella,
E.J.Goldsmith,
and
M.A.Phillips
(2007).
X-ray structure of Paramecium bursaria Chlorella virus arginine decarboxylase: insight into the structural basis for substrate specificity.
|
| |
Biochemistry,
46,
2831-2841.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Jantaro,
H.Kidron,
D.Chesnel,
A.Incharoensakdi,
P.Mulo,
T.Salminen,
and
P.Mäenpää
(2006).
Structural modeling and environmental regulation of arginine decarboxylase in Synechocystis sp. PCC 6803.
|
| |
Arch Microbiol,
184,
397-406.
|
 |
|
|
|
|
 |
R.Shah,
C.S.Coleman,
K.Mir,
J.Baldwin,
J.L.Van Etten,
N.V.Grishin,
A.E.Pegg,
B.A.Stanley,
and
M.A.Phillips
(2004).
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of eukaryotic ornithine decarboxylases.
|
| |
J Biol Chem,
279,
35760-35767.
|
 |
|
|
|
|
 |
C.V.Smith,
and
J.C.Sacchettini
(2003).
Mycobacterium tuberculosis: a model system for structural genomics.
|
| |
Curr Opin Struct Biol,
13,
658-664.
|
 |
|
|
|
|
 |
K.Gokulan,
B.Rupp,
M.S.Pavelka,
W.R.Jacobs,
and
J.C.Sacchettini
(2003).
Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis.
|
| |
J Biol Chem,
278,
18588-18596.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.K.Jackson,
E.J.Goldsmith,
and
M.A.Phillips
(2003).
X-ray structure determination of Trypanosoma brucei ornithine decarboxylase bound to D-ornithine and to G418: insights into substrate binding and ODC conformational flexibility.
|
| |
J Biol Chem,
278,
22037-22043.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.B.Balbo,
C.N.Patel,
K.G.Sell,
R.S.Adcock,
S.Neelakantan,
P.A.Crooks,
and
M.A.Oliveira
(2003).
Spectrophotometric and steady-state kinetic analysis of the biosynthetic arginine decarboxylase of Yersinia pestis utilizing arginine analogues as inhibitors and alternative substrates.
|
| |
Biochemistry,
42,
15189-15196.
|
 |
|
|
|
|
 |
T.T.Huynh,
V.T.Huynh,
M.A.Harmon,
and
M.A.Phillips
(2003).
Gene knockdown of gamma-glutamylcysteine synthetase by RNAi in the parasitic protozoa Trypanosoma brucei demonstrates that it is an essential enzyme.
|
| |
J Biol Chem,
278,
39794-39800.
|
 |
|
|
|
|
 |
C.Momany,
V.Levdikov,
L.Blagova,
and
K.Crews
(2002).
Crystallization of diaminopimelate decarboxylase from Escherichia coli, a stereospecific D-amino-acid decarboxylase.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
549-552.
|
 |
|
|
|
|
 |
M.Bertoldi,
M.Gonsalvi,
R.Contestabile,
and
C.B.Voltattorni
(2002).
Mutation of tyrosine 332 to phenylalanine converts dopa decarboxylase into a decarboxylation-dependent oxidative deaminase.
|
| |
J Biol Chem,
277,
36357-36362.
|
 |
|
|
|
|
 |
S.S.Ray,
J.B.Bonanno,
K.R.Rajashankar,
M.G.Pinho,
G.He,
H.De Lencastre,
A.Tomasz,
and
S.K.Burley
(2002).
Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor.
|
| |
Structure,
10,
1499-1508.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Bertoldi,
S.Castellani,
and
C.Bori Voltattorni
(2001).
Mutation of residues in the coenzyme binding pocket of Dopa decarboxylase. Effects on catalytic properties.
|
| |
Eur J Biochem,
268,
2975-2981.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

