
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.6.2.1.4
- succinate--CoA ligase (GDP-forming).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Citric acid cycle
|
 |
 |
 |
 |
 |
Reaction:
|
 |
GTP + succinate + CoA = succinyl-CoA + GDP + phosphate
|
 |
 |
 |
 |
 |
 GTP
GTP
|
+
|
 succinate
succinate
|
+
|
 CoA
CoA
|
=
|
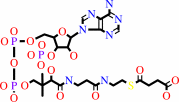 succinyl-CoA
succinyl-CoA
|
+
|
GDP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain A:
E.C.6.2.1.5
- succinate--CoA ligase (ADP-forming).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
succinate + ATP + CoA = succinyl-CoA + ADP + phosphate
|
 |
 |
 |
 |
 |
 succinate
succinate
|
+
|
 ATP
ATP
|
+
|
 CoA
CoA
|
=
|
 succinyl-CoA
succinyl-CoA
|
+
|
ADP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
299:1325-1339
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Phosphorylated and dephosphorylated structures of pig heart, GTP-specific succinyl-CoA synthetase.
|
|
M.E.Fraser,
M.N.James,
W.A.Bridger,
W.T.Wolodko.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Succinyl-CoA synthetase (SCS) catalyzes the reversible
phosphorylation/dephosphorylation reaction:¿¿¿rm succinyl ¿hbox
¿-¿CoA+NDP+P_i¿leftrightarrow succinate+CoA+NTP¿¿where N denotes adenosine
or guanosine. In the course of the reaction, an essential histidine residue is
transiently phosphorylated. We have crystallized and solved the structure of the
GTP-specific isoform of SCS from pig heart (EC 6.2.1.4) in both the
dephosphorylated and phosphorylated forms. The structures were refined to 2.1 A
resolution. In the dephosphorylated structure, the enzyme is stabilized via
coordination of a phosphate ion by the active-site histidine residue and the two
"power" helices, one contributed by each subunit of the
alphabeta-dimer. Small changes in the conformations of residues at the amino
terminus of the power helix contributed by the alpha-subunit allow the enzyme to
accommodate either the covalently bound phosphoryl group or the free phosphate
ion. Structural comparisons are made between the active sites in these two forms
of the enzyme, both of which can occur along the catalytic path. Comparisons are
also made with the structure of Escherichia coli SCS. The domain that has been
shown to bind ADP in E. coli SCS is more open in the pig heart, GTP-specific SCS
structure.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Pig heart, GTP-specific SCS. In this
three-dimensional ribbon diagram, the a-subunit is white, with
light gray for the interior of the helices, and the side-chain
of the active site phosphohistidine residue is shown as a
ball-and-stick model; the b-subunit is shaded gray, as are the
two sulfate ions. The amino and carboxy termini of each subunit
are labeled N and C, respectively. This Figure and Figure 2 and
Figure 7 were drawn using the program MOLSCRIPT (Kraulis, 1991).
|
 |
Figure 2.
Figure 2. Stereo diagrams of the regions surrounding the
active-site histidine residue. The residues are shown as
ball-and-stick models. Hydrogen-bonding interactions with the
oxygen atoms of the phosphate ion or the phosporyl group are
represented by broken lines. (a) Dephosphorylated pig heart,
GTP-specific SCS. (b) Phosphorylated pig heart, GTP-specific SCS.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2000,
299,
1325-1339)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.Tanaka,
P.Smith,
and
S.Shuman
(2010).
Structure of the RNA 3'-phosphate cyclase-adenylate intermediate illuminates nucleotide specificity and covalent nucleotidyl transfer.
|
| |
Structure,
18,
449-457.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Phillips,
A.M.Aponte,
S.A.French,
D.J.Chess,
and
R.S.Balaban
(2009).
Succinyl-CoA synthetase is a phosphate target for the activation of mitochondrial metabolism.
|
| |
Biochemistry,
48,
7140-7149.
|
 |
|
|
|
|
 |
Y.Chen,
J.Jakoncic,
K.A.Parker,
N.Carpino,
and
N.Nassar
(2009).
Structures of the phosphorylated and VO(3)-bound 2H-phosphatase domain of Sts-2.
|
| |
Biochemistry,
48,
8129-8135.
|
 |
|
|
|
|
 |
C.Bräsen,
M.Schmidt,
J.Grötzinger,
and
P.Schönheit
(2008).
Reaction mechanism and structural model of ADP-forming Acetyl-CoA synthetase from the hyperthermophilic archaeon Pyrococcus furiosus: evidence for a second active site histidine residue.
|
| |
J Biol Chem,
283,
15409-15418.
|
 |
|
|
|
|
 |
J.R.Hughes,
A.M.Meireles,
K.H.Fisher,
A.Garcia,
P.R.Antrobus,
A.Wainman,
N.Zitzmann,
C.Deane,
H.Ohkura,
and
J.G.Wakefield
(2008).
A microtubule interactome: complexes with roles in cell cycle and mitosis.
|
| |
PLoS Biol,
6,
e98.
|
 |
|
|
|
|
 |
K.Hamblin,
D.M.Standley,
M.B.Rogers,
A.Stechmann,
A.J.Roger,
R.Maytum,
and
M.van der Giezen
(2008).
Localization and nucleotide specificity of Blastocystis succinyl-CoA synthetase.
|
| |
Mol Microbiol,
68,
1395-1405.
|
 |
|
|
|
|
 |
E.Hidber,
E.R.Brownie,
K.Hayakawa,
and
M.E.Fraser
(2007).
Participation of Cys123alpha of Escherichia coli succinyl-CoA synthetase in catalysis.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
876-884.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Shikata,
T.Fukui,
H.Atomi,
and
T.Imanaka
(2007).
A novel ADP-forming succinyl-CoA synthetase in Thermococcus kodakaraensis structurally related to the archaeal nucleoside diphosphate-forming acetyl-CoA synthetases.
|
| |
J Biol Chem,
282,
26963-26970.
|
 |
|
|
|
|
 |
M.A.Joyce,
E.R.Brownie,
K.Hayakawa,
and
M.E.Fraser
(2007).
Cloning, expression, purification, crystallization and preliminary X-ray analysis of Thermus aquaticus succinyl-CoA synthetase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
399-402.
|
 |
|
|
|
|
 |
R.G.Kibbey,
R.L.Pongratz,
A.J.Romanelli,
C.B.Wollheim,
G.W.Cline,
and
G.I.Shulman
(2007).
Mitochondrial GTP regulates glucose-stimulated insulin secretion.
|
| |
Cell Metab,
5,
253-264.
|
 |
|
|
|
|
 |
J.S.Lott,
B.Paget,
J.M.Johnston,
L.T.Delbaere,
J.A.Sigrell-Simon,
M.J.Banfield,
and
E.N.Baker
(2006).
The structure of an ancient conserved domain establishes a structural basis for stable histidine phosphorylation and identifies a new family of adenosine-specific kinases.
|
| |
J Biol Chem,
281,
22131-22141.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.E.Fraser,
K.Hayakawa,
M.S.Hume,
D.G.Ryan,
and
E.R.Brownie
(2006).
Interactions of GTP with the ATP-grasp domain of GTP-specific succinyl-CoA synthetase.
|
| |
J Biol Chem,
281,
11058-11065.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.D.Busam,
A.G.Thorsell,
A.Flores,
M.Hammarström,
C.Persson,
and
B.M.Hallberg
(2006).
First structure of a eukaryotic phosphohistidine phosphatase.
|
| |
J Biol Chem,
281,
33830-33834.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Kothe,
and
S.G.Powers-Lee
(2004).
Nucleotide recognition in the ATP-grasp protein carbamoyl phosphate synthetase.
|
| |
Protein Sci,
13,
466-475.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

