
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain A:
E.C.1.3.1.14
- dihydroorotate dehydrogenase (NAD(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-dihydroorotate + NAD+ = orotate + NADH + H+
|
 |
 |
 |
 |
 |
(S)-dihydroorotate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NAD(+)
NAD(+)
|
=
|
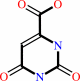 orotate
orotate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; FMN; Iron-sulfur
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
FMN
Bound ligand (Het Group name =
FMN)
corresponds exactly
|
 Iron-sulfur
Iron-sulfur
|
|
 |
 |
Enzyme class 3:
|
 |
Chain B:
E.C.1.3.3.1
- Transferred entry: 1.3.98.1.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-dihydroorotate + O2 = orotate + H2O2
|
 |
 |
 |
 |
 |
(S)-dihydroorotate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 O(2)
O(2)
|
=
|
 orotate
orotate
|
+
|
 H(2)O(2)
H(2)O(2)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; FMN
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
FMN
Bound ligand (Het Group name =
FMN)
corresponds exactly
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
8:1227-1238
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
|
|
P.Rowland,
S.Nørager,
K.F.Jensen,
S.Larsen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: The fourth step and only redox reaction in pyrimidine de novo
biosynthesis is catalyzed by the flavoprotein dihydroorotate dehydrogenase
(DHOD). Based on their sequences, DHODs are grouped into two major families.
Lactococcus lactis is one of the few organisms with two DHODs, A and B,
belonging to each of the two subgroups of family 1. The B enzyme (DHODB) is a
prototype for DHODs in Gram-positive bacteria that use NAD+ as the second
substrate. DHODB is a heterotetramer composed of two different proteins (PyrDB
and PyrK) and three different cofactors: FMN, FAD, and a [2Fe-2S] cluster.
RESULTS: Crystal structures have been determined for DHODB and its product
complex. The DHODB heterotetramer is composed of two closely interacting
PyrDB-PyrK dimers with the [2Fe-2S] cluster in their interface centered between
the FMN and FAD groups. Conformational changes are observed between the
complexed and uncomplexed state of the enzyme for the loop carrying the
catalytic cysteine residue and one of the lysines interacting with FMN, which is
important for substrate binding. CONCLUSIONS: A dimer of two PyrDB subunits
resembling the family 1A enzymes forms the central core of DHODB. PyrK belongs
to the NADPH ferredoxin reductase superfamily. The binding site for NAD+ has
been deduced from the similarity to these proteins. The orotate binding in DHODB
is similar to that in the family 1A enzymes. The close proximity of the three
redox centers makes it possible to propose a possible electron transfer pathway
involving residues conserved among the family 1B DHODs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 6.
Figure 6. The Environment of the Two Flavin Groups and the
Orotate Binding Site in DHODB(a) The environment of the FMN
group in the uncomplexed PyrDB subunit with an open catalytic
loop. Water molecules are shown as cyan spheres.(b) The
DHODB-orotate complex structure showing the same view of the FMN
group as in Figure 6a and containing a closed but slightly
disordered catalytic loop.(c) A closeup view of the environment
of the bound orotate in the complexed structure.(d) The
environment of the FAD group in PyrK subunit in the uncomplexed
structure. No significant changes were observed in the structure
of the orotate complex

|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2000,
8,
1227-1238)
copyright 2000.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.A.Phillips,
and
P.K.Rathod
(2010).
Plasmodium dihydroorotate dehydrogenase: a promising target for novel anti-malarial chemotherapy.
|
| |
Infect Disord Drug Targets,
10,
226-239.
|
 |
|
|
|
|
 |
R.L.Fagan,
and
B.A.Palfey
(2009).
Roles in binding and chemistry for conserved active site residues in the class 2 dihydroorotate dehydrogenase from Escherichia coli.
|
| |
Biochemistry,
48,
7169-7178.
|
 |
|
|
|
|
 |
R.L.Kow,
J.R.Whicher,
C.A.McDonald,
B.A.Palfey,
and
R.L.Fagan
(2009).
Disruption of the proton relay network in the class 2 dihydroorotate dehydrogenase from Escherichia coli.
|
| |
Biochemistry,
48,
9801-9809.
|
 |
|
|
|
|
 |
S.Kawasaki,
T.Satoh,
M.Todoroki,
and
Y.Niimura
(2009).
b-type dihydroorotate dehydrogenase is purified as a H2O2-forming NADH oxidase from Bifidobacterium bifidum.
|
| |
Appl Environ Microbiol,
75,
629-636.
|
 |
|
|
|
|
 |
T.L.Arakaki,
F.S.Buckner,
J.R.Gillespie,
N.A.Malmquist,
M.A.Phillips,
O.Kalyuzhniy,
J.R.Luft,
G.T.Detitta,
C.L.Verlinde,
W.C.Van Voorhis,
W.G.Hol,
and
E.A.Merritt
(2008).
Characterization of Trypanosoma brucei dihydroorotate dehydrogenase as a possible drug target; structural, kinetic and RNAi studies.
|
| |
Mol Microbiol,
68,
37-50.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.L.Chung,
W.Wang,
and
P.E.Bourne
(2007).
High-throughput identification of interacting protein-protein binding sites.
|
| |
BMC Bioinformatics,
8,
223.
|
 |
|
|
|
|
 |
N.A.Malmquist,
J.Baldwin,
and
M.A.Phillips
(2007).
Detergent-dependent kinetics of truncated Plasmodium falciparum dihydroorotate dehydrogenase.
|
| |
J Biol Chem,
282,
12678-12686.
|
 |
|
|
|
|
 |
J.P.Combe,
J.Basran,
P.Hothi,
D.Leys,
S.E.Rigby,
A.W.Munro,
and
N.S.Scrutton
(2006).
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the catalytic cycle of dihydroorotate dehydrogenase B of Lactococcus lactis.
|
| |
J Biol Chem,
281,
17977-17988.
|
 |
|
|
|
|
 |
M.Kilstrup,
K.Hammer,
P.Ruhdal Jensen,
and
J.Martinussen
(2005).
Nucleotide metabolism and its control in lactic acid bacteria.
|
| |
FEMS Microbiol Rev,
29,
555-590.
|
 |
|
|
|
|
 |
M.Hansen,
J.Le Nours,
E.Johansson,
T.Antal,
A.Ullrich,
M.Löffler,
and
S.Larsen
(2004).
Inhibitor binding in a class 2 dihydroorotate dehydrogenase causes variations in the membrane-associated N-terminal domain.
|
| |
Protein Sci,
13,
1031-1042.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Nørager,
S.Arent,
O.Björnberg,
M.Ottosen,
L.Lo Leggio,
K.F.Jensen,
and
S.Larsen
(2003).
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function.
|
| |
J Biol Chem,
278,
28812-28822.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.C.Bishop,
J.Xu,
R.C.Johnson,
P.Schimmel,
and
V.de Crécy-Lagard
(2002).
Identification of the tRNA-dihydrouridine synthase family.
|
| |
J Biol Chem,
277,
25090-25095.
|
 |
|
|
|
|
 |
M.B.Ottosen,
O.Björnberg,
S.Nørager,
S.Larsen,
B.A.Palfey,
and
K.F.Jensen
(2002).
The dimeric dihydroorotate dehydrogenase A from Lactococcus lactis dissociates reversibly into inactive monomers.
|
| |
Protein Sci,
11,
2575-2583.
|
 |
|
|
|
|
 |
S.Nørager,
K.F.Jensen,
O.Björnberg,
and
S.Larsen
(2002).
E. coli dihydroorotate dehydrogenase reveals structural and functional distinctions between different classes of dihydroorotate dehydrogenases.
|
| |
Structure,
10,
1211-1223.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.A.Palfey,
O.Björnberg,
and
K.F.Jensen
(2001).
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase obtained by rapid reaction studies.
|
| |
Biochemistry,
40,
4381-4390.
|
 |
|
|
|
|
 |
G.Pujadas,
and
J.Palau
(2001).
Molecular mimicry of substrate oxygen atoms by water molecules in the beta-amylase active site.
|
| |
Protein Sci,
10,
1645-1657.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

