 |
PDBsum entry 1e7y

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1e7y
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.363
- glucose-6-phosphate dehydrogenase [NAD(P)(+)].
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
D-glucose 6-phosphate + NADP+ = 6-phospho-D-glucono-1,5-lactone + NADPH + H+
|
|
2.
|
D-glucose 6-phosphate + NAD+ = 6-phospho-D-glucono-1,5-lactone + NADH + H+
|
|
 |
 |
 |
 |
 |
D-glucose 6-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NADP(+)
Bound ligand (Het Group name = )
matches with 56.25% similarity
|
=
|
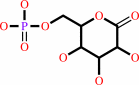 6-phospho-D-glucono-1,5-lactone
6-phospho-D-glucono-1,5-lactone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
D-glucose 6-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 47.92% similarity
|
=
|
 6-phospho-D-glucono-1,5-lactone
6-phospho-D-glucono-1,5-lactone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
39:15002-15011
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
An examination of the role of asp-177 in the His-Asp catalytic dyad of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase: X-ray structure and pH dependence of kinetic parameters of the D177N mutant enzyme.
|
|
M.S.Cosgrove,
S.Gover,
C.E.Naylor,
L.Vandeputte-Rutten,
M.J.Adams,
H.R.Levy.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The role of Asp-177 in the His-Asp catalytic dyad of glucose 6-phosphate
dehydrogenase from Leuconostoc mesenteroides has been investigated by a
structural and functional characterization of the D177N mutant enzyme. Its
three-dimensional structure has been determined by X-ray cryocrystallography in
the presence of NAD(+) and in the presence of glucose 6-phosphate plus NADPH.
The structure of a glucose 6-phosphate complex of a mutant (Q365C) with normal
enzyme activity has also been determined and substrate binding compared. To
understand the effect of Asp-177 on the ionization properties of the catalytic
base His-240, the pH dependence of kinetic parameters has been determined for
the D177N mutant and compared to that of the wild-type enzyme. The structures
give details of glucose 6-phosphate binding and show that replacement of the
Asp-177 of the catalytic dyad with asparagine does not affect the overall
structure of glucose 6-phosphate dehydrogenase. Additionally, the evidence
suggests that the productive tautomer of His-240 in the D177N mutant enzyme is
stabilized by a hydrogen bond with Asn-177; hence, the mutation does not affect
tautomer stabilization. We conclude, therefore, that the absence of a negatively
charged aspartate at 177 accounts for the decrease in catalytic activity at pH
7.8. Structural analysis suggests that the pH dependence of the kinetic
parameters of D177N glucose 6-phosphate dehydrogenase results from an ionized
water molecule replacing the missing negative charge of the mutated Asp-177 at
high pH. Glucose 6-phosphate binding orders and orients His-178 in the
D177N-glucose 6-phosphate-NADPH ternary complex and appears to be necessary to
form this water-binding site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Watanabe,
T.Kodaki,
T.Kodak,
and
K.Makino
(2006).
Cloning, expression, and characterization of bacterial L-arabinose 1-dehydrogenase involved in an alternative pathway of L-arabinose metabolism.
|
| |
J Biol Chem,
281,
2612-2623.
|
 |
|
|
|
|
 |
E.Ganea,
and
J.J.Harding
(2005).
Trehalose and 6-aminohexanoic acid stabilize and renature glucose-6-phosphate dehydrogenase inactivated by glycation and by guanidinium hydrochloride.
|
| |
Biol Chem,
386,
269-278.
|
 |
|
|
|
|
 |
J.Merritt,
J.A.Butz,
B.A.Ogunnaike,
and
J.S.Edwards
(2005).
Parallel analysis of mutant human glucose 6-phosphate dehydrogenase in yeast using PCR colonies.
|
| |
Biotechnol Bioeng,
92,
519-531.
|
 |
|
|
|
|
 |
M.Kotaka,
S.Gover,
L.Vandeputte-Rutten,
S.W.Au,
V.M.Lam,
and
M.J.Adams
(2005).
Structural studies of glucose-6-phosphate and NADP+ binding to human glucose-6-phosphate dehydrogenase.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
495-504.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.L.Brosius,
and
R.F.Colman
(2002).
Three subunits contribute amino acids to the active site of tetrameric adenylosuccinate lyase: Lys268 and Glu275 are required.
|
| |
Biochemistry,
41,
2217-2226.
|
 |
|
|
|
|
 |
C.E.Naylor,
S.Gover,
A.K.Basak,
M.S.Cosgrove,
H.R.Levy,
and
M.J.Adams
(2001).
NADP+ and NAD+ binding to the dual coenzyme specific enzyme Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase: different interdomain hinge angles are seen in different binary and ternary complexes.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
635-648.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

