 |
PDBsum entry 1duh
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Crystal structure of the ffh and ef-G binding sites in the conserved domain IV of escherichia coli 4.5s RNA.
|
 |
|
Authors
|
 |
L.Jovine,
T.Hainzl,
C.Oubridge,
W.G.Scott,
J.Li,
T.K.Sixma,
A.Wonacott,
T.Skarzynski,
K.Nagai.
|
 |
|
Ref.
|
 |
Structure, 2000,
8,
527-540.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
BACKGROUND: Bacterial signal recognition particle (SRP), consisting of 4.5S RNA
and Ffh protein, plays an essential role in targeting signal-peptide-containing
proteins to the secretory apparatus in the cell membrane. The 4.5S RNA increases
the affinity of Ffh for signal peptides and is essential for the interaction
between SRP and its receptor, protein FtsY. The 4.5S RNA also interacts with
elongation factor G (EF-G) in the ribosome and this interaction is required for
efficient translation. RESULTS: We have determined by multiple anomalous
dispersion (MAD) with Lu(3+) the 2.7 A crystal structure of a 4.5S RNA fragment
containing binding sites for both Ffh and EF-G. This fragment consists of three
helices connected by a symmetric and an asymmetric internal loop. In contrast to
NMR-derived structures reported previously, the symmetric loop is entirely
constituted by non-canonical base pairs. These pairs continuously stack and
project unusual sets of hydrogen-bond donors and acceptors into the shallow
minor groove. The structure can therefore be regarded as two double helical rods
hinged by the asymmetric loop that protrudes from one strand. CONCLUSIONS: Based
on our crystal structure and results of chemical protection experiments reported
previously, we predicted that Ffh binds to the minor groove of the symmetric
loop. An identical decanucleotide sequence is found in the EF-G binding sites of
both 4.5S RNA and 23S rRNA. The decanucleotide structure in the 4.5S RNA and the
ribosomal protein L11-RNA complex crystals suggests how 4.5S RNA and 23S rRNA
might interact with EF-G and function in translating ribosomes.
|
 |
 |
 |

|
 |
Figure 2.
Figure 2. Stereo ball-and-stick representation of the
symmetric loop A region of the refined 45 RNA crystal structure,
with combined, sigmaa-weighted |2F[o]-F[c]| electron-density map
contoured at 1.0s.
|
 |
|
 |
 |
|
The above figure is
reprinted
by permission from Cell Press:
Structure
(2000,
8,
527-540)
copyright 2000.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
Crystallization and preliminary X-Ray analysis of the conserved domain IV of escherichia coli 4.5s RNA.
|
 |
|
Authors
|
 |
L.Jovine,
T.Hainz,
C.Oubridge,
K.Nagai.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2000,
56,
1033-1037.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |
Figure 2.
Figure 2 Predicted folding of a generic 5' hammerhead
ribozyme-SRP RNA-3' hepatitis  virus
ribozyme transcript (in this case, the sequence of the 45 RNA
construct is shown in red). Bases of the transcription plasmid
insert important for efficient T7 RNA polymerase transcription
and ribozyme activity are boxed in white and black,
respectively. Cloning sites are in brackets and 5' and 3'
ribozyme cleavage sites are indicated by black arrows.
Constructs of this type were cloned into the XbaI/PstI sites of
a pUC18-based vector carrying a 5' T7 promoter and a 3' fragment
of an hepatitis virus
ribozyme transcript (in this case, the sequence of the 45 RNA
construct is shown in red). Bases of the transcription plasmid
insert important for efficient T7 RNA polymerase transcription
and ribozyme activity are boxed in white and black,
respectively. Cloning sites are in brackets and 5' and 3'
ribozyme cleavage sites are indicated by black arrows.
Constructs of this type were cloned into the XbaI/PstI sites of
a pUC18-based vector carrying a 5' T7 promoter and a 3' fragment
of an hepatitis  virus
ribozyme sequence (pUC18T7Pst virus
ribozyme sequence (pUC18T7Pst  V;
Dr Sandra Searles, MRC-LMB, personal communication). This
created a new 5' hammerhead ribozyme sequence and reconstituted
a complete 3' hepatitis V;
Dr Sandra Searles, MRC-LMB, personal communication). This
created a new 5' hammerhead ribozyme sequence and reconstituted
a complete 3' hepatitis  virus
ribozyme sequence. After digestion of the resulting plasmid at
the 3' HindIII site (indicated by a blue arrow) and in vitro
run-off transcription/co-transcriptional RNA 5'-end cleavage by
hammerhead ribozyme (Price, Oubridge et al., 1998[Price, S. R.,
Oubridge, C., Varani, G. & Nagai, K. (1998). RNA-Protein
Interactions: A Practical Approach, edited by C. Smith, pp.
37-74. Oxford University Press.]), 3'-end cleavage of the RNA
product by the hepatitis virus
ribozyme sequence. After digestion of the resulting plasmid at
the 3' HindIII site (indicated by a blue arrow) and in vitro
run-off transcription/co-transcriptional RNA 5'-end cleavage by
hammerhead ribozyme (Price, Oubridge et al., 1998[Price, S. R.,
Oubridge, C., Varani, G. & Nagai, K. (1998). RNA-Protein
Interactions: A Practical Approach, edited by C. Smith, pp.
37-74. Oxford University Press.]), 3'-end cleavage of the RNA
product by the hepatitis 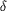 virus
ribozyme required a 2 min annealing step at 338 K followed by
a 4 h incubation at 328 K. virus
ribozyme required a 2 min annealing step at 338 K followed by
a 4 h incubation at 328 K.
|
 |
|
 |
 |
|
The above figure is
reproduced from the cited reference
with permission from the IUCr
|
 |
|

|

|
|
|
Headers
|
 |
|
|
|
 |

