 |
PDBsum entry 1ddi

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ddi
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.1.2
- assimilatory sulfite reductase (NADPH).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hydrogen sulfide + 3 NADP+ + 3 H2O = sulfite + 3 NADPH + 4 H+
|
 |
 |
 |
 |
 |
hydrogen sulfide
|
+
|
 3
×
NADP(+)
3
×
NADP(+)
|
+
|
3
×
H2O
|
=
|
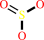 sulfite
sulfite
|
+
|
 3
×
NADPH
3
×
NADPH
|
+
|
4
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
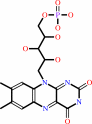
FAD; FMN; Iron-sulfur; Siroheme
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
 FMN
FMN
|
 Iron-sulfur
Iron-sulfur
|
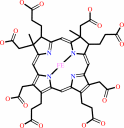 Siroheme
Siroheme
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
299:199-212
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
|
|
A.Gruez,
D.Pignol,
M.Zeghouf,
J.Covès,
M.Fontecave,
J.L.Ferrer,
J.C.Fontecilla-Camps.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Escherichia coli NADPH-sulfite reductase (SiR) is a 780 kDa multimeric
hemoflavoprotein composed of eight alpha-subunits (SiR-FP) and four
beta-subunits (SiR-HP) that catalyses the six electron reduction of sulfite to
sulfide. Each beta-subunit contains a Fe4S4 cluster and a siroheme, and each
alpha-subunit binds one FAD and one FMN as prosthetic groups. The FAD gets
electrons from NADPH, and the FMN transfers the electrons to the metal centers
of the beta-subunit for sulfite reduction. We report here the 1.94 A X-ray
structure of SiR-FP60, a recombinant monomeric fragment of SiR-FP that binds
both FAD and FMN and retains the catalytic properties of the native protein. The
structure can be divided into three domains. The carboxy-terminal part of the
enzyme is composed of an antiparallel beta-barrel which binds the FAD, and a
variant of the classical pyridine dinucleotide binding fold which binds NADPH.
These two domains form the canonic FNR-like module, typical of the ferredoxin
NADP+ reductase family. By analogy with the structure of the cytochrome P450
reductase, the third domain, composed of seven alpha-helices, is supposed to
connect the FNR-like module to the N-terminal flavodoxine-like module. In four
different crystal forms, the FMN-binding module is absent from electron density
maps, although mass spectroscopy, amino acid sequencing and activity experiments
carried out on dissolved crystals indicate that a functional module is present
in the protein. Our results clearly indicate that the interaction between the
FNR-like and the FMN-like modules displays lower affinity than in the case of
cytochrome P450 reductase. The flexibility of the FMN-binding domain may be
related, as observed in the case of cytochrome bc1, to a domain reorganisation
in the course of electron transfer. Thus, a movement of the FMN-binding domain
relative to the rest of the enzyme may be a requirement for its optimal
positioning relative to both the FNR-like module and the beta-subunit.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Ribbon diagram of SiR-
FP60 produced with the program
MOLSCRIPT (Kraulis, 1991) and
Render 3D (Merrit & Bacon, 1997).
The colors vary from blue for the
N-terminal residue to red for the
C-terminal residue. Cofactors are
represented by balls and sticks.
|
 |
Figure 7.
Figure 7. Electrostatic potential surfaces of SiR-FP60 and cytochrome P450 reductase (CPR). (a) The relative orien-
tations of the FMN-binding domain (gold) and the FNR-like module (silver) in CPR. The electrostatic potential sur-
faces of the FNR-like module of CPR (b) and of SiR-FP60 (c) are depicted in the same orientation as in (a). The
complementary surface of the FMN-binding domain of CPR (d) and of a model of the equivalent region of SiR-FP60
(e) are shown in the side opposite to (a), obtained by 180 ° rotation about the y-axis. SiR-FP60 FMN-binding domain
was modeled using the CPR coordinates,with the program Modeller (Sali & Blundell, 1993). The Figure was prepared
with the program Grasp (Nicholls et al., 1991).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2000,
299,
199-212)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Aigrain,
D.Pompon,
and
G.Truan
(2011).
Role of the interface between the FMN and FAD domains in the control of redox potential and electronic transfer of NADPH-cytochrome P450 reductase.
|
| |
Biochem J,
435,
197-206.
|
 |
|
|
|
|
 |
S.E.Rigby,
X.Lou,
H.S.Toogood,
K.R.Wolthers,
and
N.S.Scrutton
(2011).
ELDOR Spectroscopy Reveals that Energy Landscapes in Human Methionine Synthase Reductase are Extensively Remodelled Following Ligand and Partner Protein Binding.
|
| |
Chembiochem,
12,
863-867.
|
 |
|
|
|
|
 |
Y.C.Hsieh,
M.Y.Liu,
V.C.Wang,
Y.L.Chiang,
E.H.Liu,
W.G.Wu,
S.I.Chan,
and
C.J.Chen
(2010).
Structural insights into the enzyme catalysis from comparison of three forms of dissimilatory sulphite reductase from Desulfovibrio gigas.
|
| |
Mol Microbiol,
78,
1101-1116.
|
 |
|
|
|
|
 |
J.Ellis,
A.Gutierrez,
I.L.Barsukov,
W.C.Huang,
J.G.Grossmann,
and
G.C.Roberts
(2009).
Domain motion in cytochrome P450 reductase: conformational equilibria revealed by NMR and small-angle x-ray scattering.
|
| |
J Biol Chem,
284,
36628-36637.
|
 |
|
|
|
|
 |
M.V.Navarro,
N.De,
N.Bae,
Q.Wang,
and
H.Sondermann
(2009).
Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX.
|
| |
Structure,
17,
1104-1116.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.A.Musumeci,
A.K.Arakaki,
D.V.Rial,
D.L.Catalano-Dupuy,
and
E.A.Ceccarelli
(2008).
Modulation of the enzymatic efficiency of ferredoxin-NADP(H) reductase by the amino acid volume around the catalytic site.
|
| |
FEBS J,
275,
1350-1366.
|
 |
|
|
|
|
 |
M.Medina,
R.Abagyan,
C.Gómez-Moreno,
and
J.Fernandez-Recio
(2008).
Docking analysis of transient complexes: interaction of ferredoxin-NADP+ reductase with ferredoxin and flavodoxin.
|
| |
Proteins,
72,
848-862.
|
 |
|
|
|
|
 |
D.W.Konas,
K.Zhu,
M.Sharma,
K.S.Aulak,
G.W.Brudvig,
and
D.J.Stuehr
(2004).
The FAD-shielding residue Phe1395 regulates neuronal nitric-oxide synthase catalysis by controlling NADP+ affinity and a conformational equilibrium within the flavoprotein domain.
|
| |
J Biol Chem,
279,
35412-35425.
|
 |
|
|
|
|
 |
E.D.Garcin,
C.M.Bruns,
S.J.Lloyd,
D.J.Hosfield,
M.Tiso,
R.Gachhui,
D.J.Stuehr,
J.A.Tainer,
and
E.D.Getzoff
(2004).
Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase.
|
| |
J Biol Chem,
279,
37918-37927.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.S.Toogood,
A.van Thiel,
J.Basran,
M.J.Sutcliffe,
N.S.Scrutton,
and
D.Leys
(2004).
Extensive domain motion and electron transfer in the human electron transferring flavoprotein.medium chain Acyl-CoA dehydrogenase complex.
|
| |
J Biol Chem,
279,
32904-32912.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Panda,
S.Adak,
D.Konas,
M.Sharma,
and
D.J.Stuehr
(2004).
A conserved aspartate (Asp-1393) regulates NADPH reduction of neuronal nitric-oxide synthase: implications for catalysis.
|
| |
J Biol Chem,
279,
18323-18333.
|
 |
|
|
|
|
 |
A.Gutierrez,
A.W.Munro,
A.Grunau,
C.R.Wolf,
N.S.Scrutton,
and
G.C.Roberts
(2003).
Interflavin electron transfer in human cytochrome P450 reductase is enhanced by coenzyme binding. Relaxation kinetic studies with coenzyme analogues.
|
| |
Eur J Biochem,
270,
2612-2621.
|
 |
|
|
|
|
 |
D.Leys,
J.Basran,
F.Talfournier,
M.J.Sutcliffe,
and
N.S.Scrutton
(2003).
Extensive conformational sampling in a ternary electron transfer complex.
|
| |
Nat Struct Biol,
10,
219-225.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.M.Knudsen,
C.R.Nishida,
S.D.Mooney,
and
P.R.Ortiz de Montellano
(2003).
Nitric-oxide synthase (NOS) reductase domain models suggest a new control element in endothelial NOS that attenuates calmodulin-dependent activity.
|
| |
J Biol Chem,
278,
31814-31824.
|
 |
|
|
|
|
 |
N.Carrillo,
and
E.A.Ceccarelli
(2003).
Open questions in ferredoxin-NADP+ reductase catalytic mechanism.
|
| |
Eur J Biochem,
270,
1900-1915.
|
 |
|
|
|
|
 |
Z.W.Guan,
D.Kamatani,
S.Kimura,
and
T.Iyanagi
(2003).
Mechanistic studies on the intramolecular one-electron transfer between the two flavins in the human neuronal nitric-oxide synthase and inducible nitric-oxide synthase flavin domains.
|
| |
J Biol Chem,
278,
30859-30868.
|
 |
|
|
|
|
 |
M.Jones,
F.Talfournier,
A.Bobrov,
J.G.Grossmann,
N.Vekshin,
M.J.Sutcliffe,
and
N.S.Scrutton
(2002).
Electron transfer and conformational change in complexes of trimethylamine dehydrogenase and electron transferring flavoprotein.
|
| |
J Biol Chem,
277,
8457-8465.
|
 |
|
|
|
|
 |
S.Adak,
M.Sharma,
A.L.Meade,
and
D.J.Stuehr
(2002).
A conserved flavin-shielding residue regulates NO synthase electron transfer and nicotinamide coenzyme specificity.
|
| |
Proc Natl Acad Sci U S A,
99,
13516-13521.
|
 |
|
|
|
|
 |
S.L.Cohen,
and
B.T.Chait
(2001).
Mass spectrometry as a tool for protein crystallography.
|
| |
Annu Rev Biophys Biomol Struct,
30,
67-85.
|
 |
|
|
|
|
 |
A.Gutierrez,
O.Doehr,
M.Paine,
C.R.Wolf,
N.S.Scrutton,
and
G.C.Roberts
(2000).
Trp-676 facilitates nicotinamide coenzyme exchange in the reductive half-reaction of human cytochrome P450 reductase: properties of the soluble W676H and W676A mutant reductases.
|
| |
Biochemistry,
39,
15990-15999.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

