 |
PDBsum entry 1cne

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1cne
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.7.1.1
- nitrate reductase (NADH).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
nitrite + NAD+ + H2O = nitrate + NADH + H+
|
 |
 |
 |
 |
 |
 nitrite
nitrite
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
 nitrate
nitrate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD or FMN; Heme; Mo cation
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
or
|
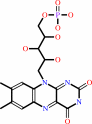 FMN
FMN
|
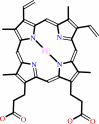 Heme
Heme
|
Mo cation
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Mol Biol
248:931-948
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
|
|
G.Lu,
Y.Lindqvist,
G.Schneider,
U.Dwivedi,
W.Campbell.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The refined crystal structures of the recombinant cytochrome b reductase
fragment of corn (Zea mays) nitrate reductase, its ADP complex and the
active-site mutant Cys242Ser are reported here. The native structure has been
refined at 2.5 A resolution to a crystallographic R-factor of 18.7% with
root-mean-square (r.m.s) deviations from standard bond lengths and angles of
0.013 A and 2.0 degrees. The diffraction pattern of the crystals is highly
anisotropic and correction of this effect lowered the crystallographic R-factor
by 5% during the refinement. The structure of the enzyme co-crystallized with
ADP has been solved at 2.7 A resolution and refined to an R-factor of 18.6% with
r.m.s. deviations from standard bond lengths and angles of 0.014 A and 2.1
degrees. It revealed the binding site of the ADP moiety of the NADH cofactor,
which is the electron donor for nitrate reduction. Based on this structure, a
model of NADH at the active site of the enzyme was built and the implications
for electron transfer from NADH to the flavin cofactor are discussed. The
crystal structure of an active-site mutant enzyme, Cys242Ser, has been solved by
difference Fourier synthesis and refined to an R-factor of 19.0% to 3.0 A
resolution with standard deviations of bond lengths and angles of 0.017 A and
2.5 degrees. This structure analysis suggests that the observed decrease in
catalytic activity of this mutant might be due to misalignment of the
nicotinamide ring in its binding site. A model of the heme-containing domain of
nitrate reductase has been built based on the X-ray structure of bovine
cytochrome b5 and has been docked with the cytochrome b reductase fragment of
nitrate reductase. The model of the complex contains six salt-bridges at the
domain-domain interface and a hydrophobic core. In this model, His48, an
invariant residue in the cytochrome b reductase family, forms an interaction
with the propionic acid group of the D-ring of the heme cofactor. This group is
in contact with the C-8 methyl group of the flavin ring. Residues that might
influence the redox potential of the flavin cofactor are proposed and their
possible role in electron transfer is discussed.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Aono,
Y.Sakamoto,
M.Miura,
F.Takeuchi,
H.Hori,
and
M.Tsubaki
(2010).
Direct electrochemical analyses of human cytochromes b5 with a mutated heme pocket showed a good correlation between their midpoint and half wave potentials.
|
| |
J Biomed Sci,
17,
90.
|
 |
|
|
|
|
 |
J.B.Glass,
F.Wolfe-Simon,
and
A.D.Anbar
(2009).
Coevolution of metal availability and nitrogen assimilation in cyanobacteria and algae.
|
| |
Geobiology,
7,
100-123.
|
 |
|
|
|
|
 |
G.J.Workun,
K.Moquin,
R.A.Rothery,
and
J.H.Weiner
(2008).
Evolutionary persistence of the molybdopyranopterin-containing sulfite oxidase protein fold.
|
| |
Microbiol Mol Biol Rev,
72,
228.
|
 |
|
|
|
|
 |
G.Schwarz,
and
R.R.Mendel
(2006).
Molybdenum cofactor biosynthesis and molybdenum enzymes.
|
| |
Annu Rev Plant Biol,
57,
623-647.
|
 |
|
|
|
|
 |
R.Kumar,
J.G.Wallis,
C.Skidmore,
and
J.Browse
(2006).
A mutation in Arabidopsis cytochrome b5 reductase identified by high-throughput screening differentially affects hydroxylation and desaturation.
|
| |
Plant J,
48,
920-932.
|
 |
|
|
|
|
 |
G.G.Barbier,
and
W.H.Campbell
(2005).
Viscosity effects on eukaryotic nitrate reductase activity.
|
| |
J Biol Chem,
280,
26049-26054.
|
 |
|
|
|
|
 |
R.R.Mendel
(2005).
Molybdenum: biological activity and metabolism.
|
| |
Dalton Trans,
(),
3404-3409.
|
 |
|
|
|
|
 |
R.Sankararamakrishnan,
S.Verma,
and
S.Kumar
(2005).
ATCUN-like metal-binding motifs in proteins: identification and characterization by crystal structure and sequence analysis.
|
| |
Proteins,
58,
211-221.
|
 |
|
|
|
|
 |
K.Oikawa,
S.Kimura,
N.Aoki,
Y.Atsuta,
Y.Takiyama,
T.Nagato,
M.Yanai,
H.Kobayashi,
K.Sato,
T.Sasajima,
and
M.Tateno
(2004).
Neuronal calcium sensor protein visinin-like protein-3 interacts with microsomal cytochrome b5 in a Ca2+-dependent manner.
|
| |
J Biol Chem,
279,
15142-15152.
|
 |
|
|
|
|
 |
K.Türk,
A.Puhar,
F.Neese,
E.Bill,
G.Fritz,
and
J.Steuber
(2004).
NADH oxidation by the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae: functional role of the NqrF subunit.
|
| |
J Biol Chem,
279,
21349-21355.
|
 |
|
|
|
|
 |
S.Bando,
T.Takano,
T.Yubisui,
K.Shirabe,
M.Takeshita,
and
A.Nakagawa
(2004).
Structure of human erythrocyte NADH-cytochrome b5 reductase.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1929-1934.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Tomiki,
and
N.Saitou
(2004).
Phylogenetic analysis of proteins associated in the four major energy metabolism systems: photosynthesis, aerobic respiration, denitrification, and sulfur respiration.
|
| |
J Mol Evol,
59,
158-176.
|
 |
|
|
|
|
 |
D.Grabowska,
D.Plochocka,
E.Jablonska-Skwiecinska,
A.Chelstowska,
I.Lewandowska,
K.Staniszewska,
Z.Majewska,
I.Witos,
and
B.Burzynska
(2003).
Compound heterozygosity of two missense mutations in the NADH-cytochrome b5 reductase gene of a Polish patient with type I recessive congenital methaemoglobinaemia.
|
| |
Eur J Haematol,
70,
404-409.
|
 |
|
|
|
|
 |
J.B.Schenkman,
and
I.Jansson
(2003).
The many roles of cytochrome b5.
|
| |
Pharmacol Ther,
97,
139-152.
|
 |
|
|
|
|
 |
R.Hille
(2002).
Molybdenum and tungsten in biology.
|
| |
Trends Biochem Sci,
27,
360-367.
|
 |
|
|
|
|
 |
H.J.Chiu,
E.Johnson,
I.Schröder,
and
D.C.Rees
(2001).
Crystal structures of a novel ferric reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus and its complex with NADP+.
|
| |
Structure,
9,
311-319.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Xoconostle-Cázares,
R.Ruiz-Medrano,
and
W.J.Lucas
(2000).
Proteolytic processing of CmPP36, a protein from the cytochrome b(5) reductase family, is required for entry into the phloem translocation pathway.
|
| |
Plant J,
24,
735-747.
|
 |
|
|
|
|
 |
J.Wu,
J.H.Gan,
Z.X.Xia,
Y.H.Wang,
W.H.Wang,
L.L.Xue,
Y.Xie,
and
Z.X.Huang
(2000).
Crystal structure of recombinant trypsin-solubilized fragment of cytochrome b(5) and the structural comparison with Val61His mutant.
|
| |
Proteins,
40,
249-257.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.L.Roberts,
D.Salazar,
J.P.Fulmer,
F.E.Frerman,
and
J.J.Kim
(1999).
Crystal structure of Paracoccus denitrificans electron transfer flavoprotein: structural and electrostatic analysis of a conserved flavin binding domain.
|
| |
Biochemistry,
38,
1977-1989.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Pattanayak,
and
S.R.Chatterjee
(1999).
Inactivation of sunflower NADH:nitrate reductase by white light-activated rose bengal.
|
| |
Mol Cell Biol Res Commun,
1,
237-240.
|
 |
|
|
|
|
 |
M.Ingelman,
S.Ramaswamy,
V.Nivière,
M.Fontecave,
and
H.Eklund
(1999).
Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli.
|
| |
Biochemistry,
38,
7040-7049.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Kimura,
Y.Emi,
S.Ikushiro,
and
T.Iyanagi
(1999).
Systematic mutations of highly conserved His49 and carboxyl-terminal of recombinant porcine liver NADH-cytochrome b5 reductase solubilized domain.
|
| |
Biochim Biophys Acta,
1430,
290-301.
|
 |
|
|
|
|
 |
W.H.Campbell
(1999).
NITRATE REDUCTASE STRUCTURE, FUNCTION AND REGULATION: Bridging the Gap between Biochemistry and Physiology.
|
| |
Annu Rev Plant Physiol Plant Mol Biol,
50,
277-303.
|
 |
|
|
|
|
 |
C.Kisker,
H.Schindelin,
D.Baas,
J.Rétey,
R.U.Meckenstock,
and
P.M.Kroneck
(1998).
A structural comparison of molybdenum cofactor-containing enzymes.
|
| |
FEMS Microbiol Rev,
22,
503-521.
|
 |
|
|
|
|
 |
G.Sridhar Prasad,
N.Kresge,
A.B.Muhlberg,
A.Shaw,
Y.S.Jung,
B.K.Burgess,
and
C.D.Stout
(1998).
The crystal structure of NADPH:ferredoxin reductase from Azotobacter vinelandii.
|
| |
Protein Sci,
7,
2541-2549.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.Nivière,
M.A.Vanoni,
G.Zanetti,
and
M.Fontecave
(1998).
Reaction of the NAD(P)H:flavin oxidoreductase from Escherichia coli with NADPH and riboflavin: identification of intermediates.
|
| |
Biochemistry,
37,
11879-11887.
|
 |
|
|
|
|
 |
C.Kisker,
H.Schindelin,
and
D.C.Rees
(1997).
Molybdenum-cofactor-containing enzymes: structure and mechanism.
|
| |
Annu Rev Biochem,
66,
233-267.
|
 |
|
|
|
|
 |
K.Ratnam,
N.Shiraishi,
W.H.Campbell,
and
R.Hille
(1997).
Spectroscopic and kinetic characterization of the recombinant cytochrome c reductase fragment of nitrate reductase. Identification of the rate-limiting catalytic step.
|
| |
J Biol Chem,
272,
2122-2128.
|
 |
|
|
|
|
 |
M.Wang,
D.L.Roberts,
R.Paschke,
T.M.Shea,
B.S.Masters,
and
J.J.Kim
(1997).
Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes.
|
| |
Proc Natl Acad Sci U S A,
94,
8411-8416.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Sakurai,
and
T.Sakurai
(1997).
Isolation and characterization of nitric oxide reductase from Paracoccus halodenitrificans.
|
| |
Biochemistry,
36,
13809-13815.
|
 |
|
|
|
|
 |
H.Nishida,
and
K.Miki
(1996).
Electrostatic properties deduced from refined structures of NADH-cytochrome b5 reductase and the other flavin-dependent reductases: pyridine nucleotide-binding and interaction with an electron-transfer partner.
|
| |
Proteins,
26,
32-41.
|
 |
|
|
|
|
 |
V.Nivière,
F.Fieschi,
J.L.Décout,
and
M.Fontecave
(1996).
Is the NAD(P)H:flavin oxidoreductase from Escherichia coli a member of the ferredoxin-NADP+ reductase family?. Evidence for the catalytic role of serine 49 residue.
|
| |
J Biol Chem,
271,
16656-16661.
|
 |
|
|
|
|
 |
K.Ratnam,
N.Shiraishi,
W.H.Campbell,
and
R.Hille
(1995).
Spectroscopic and kinetic characterization of the recombinant wild-type and C242S mutant of the cytochrome b reductase fragment of nitrate reductase.
|
| |
J Biol Chem,
270,
24067-24072.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

