 |
PDBsum entry 1bw0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.5
- tyrosine transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Phenylalanine and Tyrosine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-tyrosine + 2-oxoglutarate = 3-(4-hydroxyphenyl)pyruvate + L-glutamate
|
 |
 |
 |
 |
 |
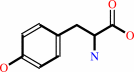 L-tyrosine
L-tyrosine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
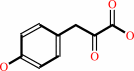 3-(4-hydroxyphenyl)pyruvate
3-(4-hydroxyphenyl)pyruvate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
8:2406-2417
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Trypanosoma cruzi tyrosine aminotransferase: substrate specificity is influenced by cofactor binding mode.
|
|
W.Blankenfeldt,
C.Nowicki,
M.Montemartini-Kalisz,
H.M.Kalisz,
H.J.Hecht.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of tyrosine aminotransferase (TAT) from the parasitic
protozoan Trypanosoma cruzi, which belongs to the aminotransferase subfamily
Igamma, has been determined at 2.5 A resolution with the R-value R = 15.1%. T.
cruzi TAT shares less than 15% sequence identity with aminotransferases of
subfamily Ialpha but shows only two larger topological differences to the
aspartate aminotransferases (AspATs). First, TAT contains a loop protruding from
the enzyme surface in the larger cofactor-binding domain, where the AspATs have
a kinked alpha-helix. Second, in the smaller substrate-binding domain, TAT has a
four-stranded antiparallel beta-sheet instead of the two-stranded beta-sheet in
the AspATs. The position of the aromatic ring of the pyridoxal-5'-phosphate
cofactor is very similar to the AspATs but the phosphate group, in contrast, is
closer to the substrate-binding site with one of its oxygen atoms pointing
toward the substrate. Differences in substrate specificities of T. cruzi TAT and
subfamily Ialpha aminotransferases can be attributed by modeling of substrate
complexes mainly to this different position of the cofactor-phosphate group.
Absence of the arginine, which in the AspATs fixes the substrate side-chain
carboxylate group by a salt bridge, contributes to the inability of T. cruzi TAT
to transaminate acidic amino acids. The preference of TAT for tyrosine is
probably related to the ability of Asn17 in TAT to form a hydrogen bond to the
tyrosine side-chain hydroxyl group.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Gonda,
E.Bar,
V.Portnoy,
S.Lev,
J.Burger,
A.A.Schaffer,
Y.Tadmor,
S.Gepstein,
J.J.Giovannoni,
N.Katzir,
and
E.Lewinsohn
(2010).
Branched-chain and aromatic amino acid catabolism into aroma volatiles in Cucumis melo L. fruit.
|
| |
J Exp Bot,
61,
1111-1123.
|
 |
|
|
|
|
 |
P.Mehere,
Q.Han,
J.A.Lemkul,
C.J.Vavricka,
H.Robinson,
D.R.Bevan,
and
J.Li
(2010).
Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations.
|
| |
Protein Cell,
1,
1023-1032.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Huang,
B.Yi,
Y.Duan,
L.Sun,
X.Yu,
J.Guo,
and
W.Chen
(2008).
Characterization and expression profiling of tyrosine aminotransferase gene from Salvia miltiorrhiza (Dan-shen) in rosmarinic acid biosynthesis pathway.
|
| |
Mol Biol Rep,
35,
601-612.
|
 |
|
|
|
|
 |
J.R.Manning,
E.R.Jefferson,
and
G.J.Barton
(2008).
The contrasting properties of conservation and correlated phylogeny in protein functional residue prediction.
|
| |
BMC Bioinformatics,
9,
51.
|
 |
|
|
|
|
 |
K.F.Stengel,
I.Holdermann,
P.Cain,
C.Robinson,
K.Wild,
and
I.Sinning
(2008).
Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
|
| |
Science,
321,
253-256.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Matsui,
and
K.Harata
(2007).
Implication for buried polar contacts and ion pairs in hyperthermostable enzymes.
|
| |
FEBS J,
274,
4012-4022.
|
 |
|
|
|
|
 |
S.Sivaraman,
and
J.F.Kirsch
(2006).
The narrow substrate specificity of human tyrosine aminotransferase--the enzyme deficient in tyrosinemia type II.
|
| |
FEBS J,
273,
1920-1929.
|
 |
|
|
|
|
 |
J.K.Yang,
C.Chang,
S.J.Cho,
J.Y.Lee,
Y.G.Yu,
S.H.Eom,
and
S.W.Suh
(2003).
Crystallization and preliminary X-ray analysis of the Mj0684 gene product, a putative aspartate aminotransferase, from Methanococcus jannaschii.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
563-565.
|
 |
|
|
|
|
 |
J.Vernal,
J.José Cazzulo,
and
C.Nowicki
(2003).
Cloning and heterologous expression of a broad specificity aminotransferase of Leishmania mexicana promastigotes1.
|
| |
FEMS Microbiol Lett,
229,
217-222.
|
 |
|
|
|
|
 |
V.R.Sobrado,
M.Montemartini-Kalisz,
H.M.Kalisz,
M.C.De La Fuente,
H.J.Hecht,
and
C.Nowicki
(2003).
Involvement of conserved asparagine and arginine residues from the N-terminal region in the catalytic mechanism of rat liver and Trypanosoma cruzi tyrosine aminotransferases.
|
| |
Protein Sci,
12,
1039-1050.
|
 |
|
|
|
|
 |
C.G.Cheong,
C.B.Bauer,
K.R.Brushaber,
J.C.Escalante-Semerena,
and
I.Rayment
(2002).
Three-dimensional structure of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica.
|
| |
Biochemistry,
41,
4798-4808.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Haruyama,
T.Nakai,
I.Miyahara,
K.Hirotsu,
H.Mizuguchi,
H.Hayashi,
and
H.Kagamiyama
(2001).
Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
|
| |
Biochemistry,
40,
4633-4644.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.I.Krupka,
R.Huber,
S.C.Holt,
and
T.Clausen
(2000).
Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
|
| |
EMBO J,
19,
3168-3178.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Feng,
M.K.Geck,
A.C.Eliot,
and
J.F.Kirsch
(2000).
Aminotransferase activity and bioinformatic analysis of 1-aminocyclopropane-1-carboxylate synthase.
|
| |
Biochemistry,
39,
15242-15249.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

