 |
PDBsum entry 1buc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1buc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.8.1
- short-chain acyl-CoA dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a short-chain 2,3-saturated fatty acyl-CoA + oxidized [electron-transfer flavoprotein] + H+ = a short-chain (2E)-enoyl-CoA + reduced [electron- transfer flavoprotein]
|
 |
 |
 |
 |
 |
Butanoyl-CoA
Bound ligand (Het Group name = )
matches with 98.15% similarity
|
+
|
electron-transfer flavoprotein
|
=
|
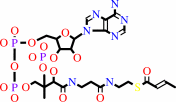 2-butenoyl-CoA
2-butenoyl-CoA
|
+
|
reduced electron-transfer flavoprotein
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
34:2163-2171
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structure of butyryl-CoA dehydrogenase from Megasphaera elsdenii.
|
|
S.Djordjevic,
C.P.Pace,
M.T.Stankovich,
J.J.Kim.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of butyryl-CoA dehydrogenase (BCAD) from Megasphaera
elsdenii complexed with acetoacetyl-CoA has been solved at 2.5 A resolution. The
enzyme crystallizes in the P422 space group with cell dimensions a = b = 107.76
A and c = 153.67 A. BCAD is a bacterial analog of short chain acyl-CoA
dehydrogenase from mammalian mitochondria. Mammalian acyl-CoA dehydrogenases are
flavin adenine dinucleotide (FAD)-containing enzymes that catalyze the first
step in the beta-oxidation of fatty acids. Although specific for substrate chain
lengths, they exhibit high sequence homology. The structure of BCAD was solved
by the molecular replacement method using the atomic coordinates of pig liver
medium chain acyl-CoA dehydrogenase (MCAD). The structure was refined to an
R-factor of 19.3%. The overall polypeptide fold of BCAD is similar to that of
MCAD. E367 in BCAD is at the same position and in a similar conformation as the
catalytic base in MCAD, E376. The main enzymatic differences between BCAD and
MCAD are their substrate specificities and the significant oxygen reactivity
exhibited by BCAD but not by MCAD. The substrate binding cavity of BCAD is
relatively shallow compared to that of MCAD, as consequences of both a single
amino acid insertion and differences in the side chains of the helices that make
the binding site. The si-face of the FAD in BCAD is more exposed to solvent than
that in MCAD. Therefore solvation can stabilize the superoxide anion and
considerably increase the rate of oxidation of reduced flavin in the bacterial
enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Z.Swigonová,
A.W.Mohsen,
and
J.Vockley
(2009).
Acyl-CoA dehydrogenases: Dynamic history of protein family evolution.
|
| |
J Mol Evol,
69,
176-193.
|
 |
|
|
|
|
 |
F.Li,
J.Hinderberger,
H.Seedorf,
J.Zhang,
W.Buckel,
and
R.K.Thauer
(2008).
Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri.
|
| |
J Bacteriol,
190,
843-850.
|
 |
|
|
|
|
 |
H.Lugg,
R.L.Sammons,
P.M.Marquis,
C.J.Hewitt,
P.Yong,
M.Paterson-Beedle,
M.D.Redwood,
A.Stamboulis,
M.Kashani,
M.Jenkins,
and
L.E.Macaskie
(2008).
Polyhydroxybutyrate accumulation by a Serratia sp.
|
| |
Biotechnol Lett,
30,
481-491.
|
 |
|
|
|
|
 |
H.Seedorf,
W.F.Fricke,
B.Veith,
H.Brüggemann,
H.Liesegang,
A.Strittmatter,
M.Miethke,
W.Buckel,
J.Hinderberger,
F.Li,
C.Hagemeier,
R.K.Thauer,
and
G.Gottschalk
(2008).
The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features.
|
| |
Proc Natl Acad Sci U S A,
105,
2128-2133.
|
 |
|
|
|
|
 |
E.S.Goetzman,
Y.Wang,
M.He,
A.W.Mohsen,
B.K.Ninness,
and
J.Vockley
(2007).
Expression and characterization of mutations in human very long-chain acyl-CoA dehydrogenase using a prokaryotic system.
|
| |
Mol Genet Metab,
91,
138-147.
|
 |
|
|
|
|
 |
J.Mackenzie,
L.Pedersen,
S.Arent,
and
A.Henriksen
(2006).
Controlling electron transfer in Acyl-CoA oxidases and dehydrogenases: a structural view.
|
| |
J Biol Chem,
281,
31012-31020.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Nomura,
M.Takada,
H.Okada,
Y.Shinohara,
T.Nakajima-Kambe,
T.Nakahara,
and
H.Uchiyama
(2005).
Identification and functional analysis of genes required for desulfurization of alkyl dibenzothiophenes of Mycobacterium sp. G3.
|
| |
J Biosci Bioeng,
100,
398-402.
|
 |
|
|
|
|
 |
R.Ensenauer,
M.He,
J.M.Willard,
E.S.Goetzman,
T.J.Corydon,
B.B.Vandahl,
A.W.Mohsen,
G.Isaya,
and
J.Vockley
(2005).
Human acyl-CoA dehydrogenase-9 plays a novel role in the mitochondrial beta-oxidation of unsaturated fatty acids.
|
| |
J Biol Chem,
280,
32309-32316.
|
 |
|
|
|
|
 |
S.Bhattacharyya,
S.Ma,
M.T.Stankovich,
D.G.Truhlar,
and
J.Gao
(2005).
Potential of mean force calculation for the proton and hydride transfer reactions catalyzed by medium-chain acyl-CoA dehydrogenase: effect of mutations on enzyme catalysis.
|
| |
Biochemistry,
44,
16549-16562.
|
 |
|
|
|
|
 |
A.Nagpal,
M.P.Valley,
P.F.Fitzpatrick,
and
A.M.Orville
(2004).
Crystallization and preliminary analysis of active nitroalkane oxidase in three crystal forms.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1456-1460.
|
 |
|
|
|
|
 |
J.J.Kim,
and
R.Miura
(2004).
Acyl-CoA dehydrogenases and acyl-CoA oxidases. Structural basis for mechanistic similarities and differences.
|
| |
Eur J Biochem,
271,
483-493.
|
 |
|
|
|
|
 |
K.P.Battaile,
T.V.Nguyen,
J.Vockley,
and
J.J.Kim
(2004).
Structures of isobutyryl-CoA dehydrogenase and enzyme-product complex: comparison with isovaleryl- and short-chain acyl-CoA dehydrogenases.
|
| |
J Biol Chem,
279,
16526-16534.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Pedersen,
and
A.Henriksen
(2004).
Expression, purification and crystallization of two peroxisomal acyl-CoA oxidases from Arabidopsis thaliana.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1125-1128.
|
 |
|
|
|
|
 |
M.Garcia-Viloca,
T.D.Poulsen,
D.G.Truhlar,
and
J.Gao
(2004).
Sensitivity of molecular dynamics simulations to the choice of the X-ray structure used to model an enzymatic reaction.
|
| |
Protein Sci,
13,
2341-2354.
|
 |
|
|
|
|
 |
M.H.Hefti,
J.Vervoort,
and
W.J.van Berkel
(2003).
Deflavination and reconstitution of flavoproteins.
|
| |
Eur J Biochem,
270,
4227-4242.
|
 |
|
|
|
|
 |
M.He,
T.P.Burghardt,
and
J.Vockley
(2003).
A novel approach to the characterization of substrate specificity in short/branched chain Acyl-CoA dehydrogenase.
|
| |
J Biol Chem,
278,
37974-37986.
|
 |
|
|
|
|
 |
K.P.Battaile,
J.Molin-Case,
R.Paschke,
M.Wang,
D.Bennett,
J.Vockley,
and
J.J.Kim
(2002).
Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases.
|
| |
J Biol Chem,
277,
12200-12207.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.V.Nguyen,
C.Riggs,
D.Babovic-Vuksanovic,
Y.S.Kim,
J.F.Carpenter,
T.P.Burghardt,
N.Gregersen,
and
J.Vockley
(2002).
Purification and characterization of two polymorphic variants of short chain acyl-CoA dehydrogenase reveal reduction of catalytic activity and stability of the Gly185Ser enzyme.
|
| |
Biochemistry,
41,
11126-11133.
|
 |
|
|
|
|
 |
C.Busquets,
B.Merinero,
E.Christensen,
J.L.Gelpí,
J.Campistol,
M.Pineda,
E.Fernández-Alvarez,
J.M.Prats,
A.Sans,
R.Arteaga,
M.Martí,
J.Campos,
M.Martínez-Pardo,
A.Martínez-Bermejo,
M.L.Ruiz-Falcó,
J.Vaquerizo,
M.Orozco,
M.Ugarte,
M.J.Coll,
and
A.Ribes
(2000).
Glutaryl-CoA dehydrogenase deficiency in Spain: evidence of two groups of patients, genetically, and biochemically distinct.
|
| |
Pediatr Res,
48,
315-322.
|
 |
|
|
|
|
 |
S.L.Volchenboum,
and
J.Vockley
(2000).
Mitochondrial import and processing of wild type and type III mutant isovaleryl-CoA dehydrogenase.
|
| |
J Biol Chem,
275,
7958-7963.
|
 |
|
|
|
|
 |
B.Nowak-Thompson,
N.Chaney,
J.S.Wing,
S.J.Gould,
and
J.E.Loper
(1999).
Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5.
|
| |
J Bacteriol,
181,
2166-2174.
|
 |
|
|
|
|
 |
T.Izard,
and
A.Geerlof
(1999).
The crystal structure of a novel bacterial adenylyltransferase reveals half of sites reactivity.
|
| |
EMBO J,
18,
2021-2030.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.J.Corydon,
P.Bross,
T.G.Jensen,
M.J.Corydon,
T.B.Lund,
U.B.Jensen,
J.J.Kim,
N.Gregersen,
and
L.Bolund
(1998).
Rapid degradation of short-chain acyl-CoA dehydrogenase variants with temperature-sensitive folding defects occurs after import into mitochondria.
|
| |
J Biol Chem,
273,
13065-13071.
|
 |
|
|
|
|
 |
S.Dakoji,
I.Shin,
K.P.Battaile,
J.Vockley,
and
H.W.Liu
(1997).
Redesigning the active-site of an acyl-CoA dehydrogenase: new evidence supporting a one-base mechanism.
|
| |
Bioorg Med Chem,
5,
2157-2164.
|
 |
|
|
|
|
 |
C.Engel,
and
R.Wierenga
(1996).
The diverse world of coenzyme A binding proteins.
|
| |
Curr Opin Struct Biol,
6,
790-797.
|
 |
|
|
|
|
 |
Z.L.Boynton,
G.N.Bennet,
and
F.B.Rudolph
(1996).
Cloning, sequencing, and expression of clustered genes encoding beta-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824.
|
| |
J Bacteriol,
178,
3015-3024.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

