 |
PDBsum entry 1b8g

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
1-aminocyclopropane-1-carboxylate synthase
|
|
Structure:
|
 |
Protein (1-aminocyclopropane-1-carboxylate synthase). Chain: a, b. Synonym: acc synthase, s-adenosyl-l-methionine methylthioadenosine- lyase. Engineered: yes
|
|
Source:
|
 |
Malus x domestica. Organism_taxid: 3750. Tissue: fruit cortical tissue. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.37Å
|
R-factor:
|
0.179
|
R-free:
|
0.242
|
|
|
Authors:
|
 |
G.Capitani,E.Hohenester,L.Feng,P.Storici,J.F.Kirsch,J.N.Jansonius
|
Key ref:

|
 |
G.Capitani
et al.
(1999).
Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene.
J Mol Biol,
294,
745-756.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
31-Jan-99
|
Release date:
|
26-Jan-00
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P37821
(1A1C_MALDO) -
1-aminocyclopropane-1-carboxylate synthase from Malus domestica
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
473 a.a.
421 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.4.-.-
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.4.1.14
- 1-aminocyclopropane-1-carboxylate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
S-adenosyl-L-methionine = 1-aminocyclopropane-1-carboxylate + S-methyl- 5'-thioadenosine + H+
|
 |
 |
 |
 |
 |
 S-adenosyl-L-methionine
S-adenosyl-L-methionine
|
=
|
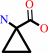 1-aminocyclopropane-1-carboxylate
1-aminocyclopropane-1-carboxylate
|
+
|
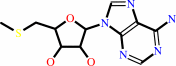 S-methyl- 5'-thioadenosine
S-methyl- 5'-thioadenosine
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
294:745-756
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene.
|
|
G.Capitani,
E.Hohenester,
L.Feng,
P.Storici,
J.F.Kirsch,
J.N.Jansonius.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The 2.4 A crystal structure of the vitamin B6-dependent enzyme
1-aminocyclopropane-1-carboxylate (ACC) synthase is described. This enzyme
catalyses the committed step in the biosynthesis of ethylene, a plant hormone
that is responsible for the initiation of fruit ripening and for regulating many
other developmental processes. ACC synthase has 15 % sequence identity with the
well-studied aspartate aminotransferase, and a completely different catalytic
activity yet the overall folds and the active sites are very similar. The new
structure together with available biochemical data enables a comparative
mechanistic analysis that largely explains the catalytic roles of the conserved
and non-conserved active site residues. An external aldimine reaction
intermediate (external aldimine with ACC, i.e. with the product) has been
modeled. The new structure provides a basis for the rational design of
inhibitors with broad agricultural applications.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Active site views of ACC synthase. (a) Stereo view of the ACC synthase active site with the most import-
ant residues labeled (prepared with the program O (Jones & Kjeldgaard, 1991)). Residues within a sphere of 11 Å of
the cofactor are depicted in yellow and atom colors, and shown in each two alternative conformation. Tyr85, which
is contributed by the neighboring subunit, is labeled with an asterisk. (b) Detailed stereo view of the ACC synthase
active site (subunit B), showing the covalent linkage between the cofactor and Lys273. The final 2Fo
-
Fc map, con-
toured at 1.2s, is superimposed on the atomic model. The model is depicted in yellow and atom colors. The unde-
fined additional density in front of the cofactor and modeled as three water molecules (red spheres) is visible at the
top right. Prepared with O.
|
 |
Figure 4.
Figure 4. Stereo view of the superposition of the ACC synthase and AATase active sites. Residues within 11 Å of
the cofactors are shown. ACC synthase is depicted in yellow, AATase in green. Residue labels for ACC synthase
appear in gold, those for AATase residues in black. Tyr85 (ACC synthase) and Tyr70 and Arg292 (AATase) are con-
tributed by the neighboring subunit and are labeled with an asterisk. Prepared with O.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1999,
294,
745-756)
copyright 1999.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.R.Choudhury,
S.K.Singh,
S.Roy,
and
D.N.Sengupta
(2010).
An insight into the sequential, structural and phylogenetic properties of banana 1-aminocyclopropane-1-carboxylate synthase 1 and study of its interaction with pyridoxal-5'-phosphate and aminoethoxyvinylglycine.
|
| |
J Biosci,
35,
281-294.
|
 |
|
|
|
|
 |
A.Tsuchisaka,
G.Yu,
H.Jin,
J.M.Alonso,
J.R.Ecker,
X.Zhang,
S.Gao,
and
A.Theologis
(2009).
A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana.
|
| |
Genetics,
183,
979.
|
 |
|
|
|
|
 |
Z.Lin,
S.Zhong,
and
D.Grierson
(2009).
Recent advances in ethylene research.
|
| |
J Exp Bot,
60,
3311-3336.
|
 |
|
|
|
|
 |
I.El-Sharkawy,
W.S.Kim,
S.Jayasankar,
A.M.Svircev,
and
D.C.Brown
(2008).
Differential regulation of four members of the ACC synthase gene family in plum.
|
| |
J Exp Bot,
59,
2009-2027.
|
 |
|
|
|
|
 |
R.Pedreschi,
E.Vanstreels,
S.Carpentier,
M.Hertog,
J.Lammertyn,
J.Robben,
J.P.Noben,
R.Swennen,
J.Vanderleyden,
and
B.M.Nicolaï
(2007).
Proteomic analysis of core breakdown disorder in Conference pears (Pyrus communis L.).
|
| |
Proteomics,
7,
2083-2099.
|
 |
|
|
|
|
 |
S.Sivaraman,
and
J.F.Kirsch
(2006).
The narrow substrate specificity of human tyrosine aminotransferase--the enzyme deficient in tyrosinemia type II.
|
| |
FEBS J,
273,
1920-1929.
|
 |
|
|
|
|
 |
H.S.Chae,
and
J.J.Kieber
(2005).
Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis.
|
| |
Trends Plant Sci,
10,
291-296.
|
 |
|
|
|
|
 |
A.Tsuchisaka,
and
A.Theologis
(2004).
Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family.
|
| |
Proc Natl Acad Sci U S A,
101,
2275-2280.
|
 |
|
|
|
|
 |
K.L.Wang,
H.Yoshida,
C.Lurin,
and
J.R.Ecker
(2004).
Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein.
|
| |
Nature,
428,
945-950.
|
 |
|
|
|
|
 |
Z.Zhang,
J.S.Ren,
I.J.Clifton,
and
C.J.Schofield
(2004).
Crystal structure and mechanistic implications of 1-aminocyclopropane-1-carboxylic acid oxidase--the ethylene-forming enzyme.
|
| |
Chem Biol,
11,
1383-1394.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Yamagami,
A.Tsuchisaka,
K.Yamada,
W.F.Haddon,
L.A.Harden,
and
A.Theologis
(2003).
Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family.
|
| |
J Biol Chem,
278,
49102-49112.
|
 |
|
|
|
|
 |
C.G.Cheong,
C.B.Bauer,
K.R.Brushaber,
J.C.Escalante-Semerena,
and
I.Rayment
(2002).
Three-dimensional structure of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica.
|
| |
Biochemistry,
41,
4798-4808.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.G.Cheong,
J.C.Escalante-Semerena,
and
I.Rayment
(2002).
Structural studies of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica: the apo, substrate, and product-aldimine complexes.
|
| |
Biochemistry,
41,
9079-9089.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.B.Kuettner,
R.Hilgenfeld,
and
M.S.Weiss
(2002).
The active principle of garlic at atomic resolution.
|
| |
J Biol Chem,
277,
46402-46407.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Capitani,
D.L.McCarthy,
H.Gut,
M.G.Grütter,
and
J.F.Kirsch
(2002).
Apple 1-aminocyclopropane-1-carboxylate synthase in complex with the inhibitor L-aminoethoxyvinylglycine. Evidence for a ketimine intermediate.
|
| |
J Biol Chem,
277,
49735-49742.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Wang,
J.B.Anderson,
J.Chen,
L.Y.Geer,
S.He,
D.I.Hurwitz,
C.A.Liebert,
T.Madej,
G.H.Marchler,
A.Marchler-Bauer,
A.R.Panchenko,
B.A.Shoemaker,
J.S.Song,
P.A.Thiessen,
R.A.Yamashita,
and
S.H.Bryant
(2002).
MMDB: Entrez's 3D-structure database.
|
| |
Nucleic Acids Res,
30,
249-252.
|
 |
|
|
|
|
 |
K.Haruyama,
T.Nakai,
I.Miyahara,
K.Hirotsu,
H.Mizuguchi,
H.Hayashi,
and
H.Kagamiyama
(2001).
Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
|
| |
Biochemistry,
40,
4633-4644.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Kakuta,
T.Igarashi,
T.Murakami,
H.Ito,
H.Matsui,
and
M.Honma
(2001).
1-Aminocyclopropane-1-carboxylate synthase of Penicillium citrinum: primary structure and expression in Escherichia coli and Saccharomyces cerevisiae.
|
| |
Biosci Biotechnol Biochem,
65,
1511-1518.
|
 |
|
|
|
|
 |
A.B.Bleecker,
and
H.Kende
(2000).
Ethylene: a gaseous signal molecule in plants.
|
| |
Annu Rev Cell Dev Biol,
16,
1.
|
 |
|
|
|
|
 |
L.Feng,
M.K.Geck,
A.C.Eliot,
and
J.F.Kirsch
(2000).
Aminotransferase activity and bioinformatic analysis of 1-aminocyclopropane-1-carboxylate synthase.
|
| |
Biochemistry,
39,
15242-15249.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

