 |
PDBsum entry 1aog

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1aog
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.1.12
- trypanothione-disulfide reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
trypanothione + NADP+ = trypanothione disulfide + NADPH + H+
|
 |
 |
 |
 |
 |
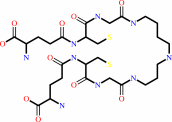 trypanothione
trypanothione
|
+
|
 NADP(+)
NADP(+)
|
=
|
 trypanothione disulfide
trypanothione disulfide
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
5:52-61
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of trypanothione reductase from the human pathogen Trypanosoma cruzi at 2.3 A resolution.
|
|
Y.Zhang,
C.S.Bond,
S.Bailey,
M.L.Cunningham,
A.H.Fairlamb,
W.N.Hunter.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Trypanothione reductase (TR) is an NADPH-dependent flavoprotein unique to
protozoan parasites from the genera Trypanosoma and Leishmania and is an
important target for the design of improved trypanocidal drugs. We present
details of the structure of TR from the human pathogen Trypanosoma cruzi, the
agent responsible for Chagas' disease or South American trypanosomiasis. The
structure has been solved by molecular replacement, using as the starting model
the structure of the enzyme from the nonpathogenic Crithidia fasciculata, and
refined to an R-factor of 18.9% for 53,868 reflections with F > or = sigma F
between 8.0 and 2.3 A resolution. The model comprises two subunits (968
residues), two FAD prosthetic groups, two maleate ions, and 419 water molecules.
The accuracy and geometry of the enzyme model is improved with respect to the C.
fasciculata enzyme model. The new structure is described and specific features
of the enzyme involved in substrate interactions are compared with previous
models of TR and related glutathione reductases from human and Escherichia coli.
Structural differences at the edge of the active sites suggest an explanation
for the differing specificities toward glutathionylspermidine disulfide.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 7.
Fig. 7. Distribution of chrgeinandaroundthe disulfide bindingsites
of (A) T. cruzi TR; (B) human GR; and (C) E. oli GR. Figurespro-
ducedwith (Nicholls, 1993) and coloredas follows: white,neu-
tral; red, negative harge; blue, positive charges. Coordinates for GR
structureswereretrievd from the Brookhaven Protein Data Bank.
|
 |
Figure 10.
Fig. 10. Stereo view t show thetriad Glu-His-Water system in active site Athat may protonation of th active site base
His 461. Blue dashed lines represent hydrogen bonds.Atoms are colored as follows: N, cyan; 0, red; C, grey.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the Protein Society:
Protein Sci
(1996,
5,
52-61)
copyright 1996.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.C.Jones,
A.Ariza,
W.H.Chow,
S.L.Oza,
and
A.H.Fairlamb
(2010).
Comparative structural, kinetic and inhibitor studies of Trypanosoma brucei trypanothione reductase with T. cruzi.
|
| |
Mol Biochem Parasitol,
169,
12-19.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.K.Fyfe,
M.S.Alphey,
and
W.N.Hunter
(2010).
Structure of Trypanosoma brucei glutathione synthetase: domain and loop alterations in the catalytic cycle of a highly conserved enzyme.
|
| |
Mol Biochem Parasitol,
170,
93-99.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Patel,
S.Hussain,
R.Harris,
S.Sardiwal,
J.M.Kelly,
S.R.Wilkinson,
P.C.Driscoll,
and
S.Djordjevic
(2010).
Structural insights into the catalytic mechanism of Trypanosoma cruzi GPXI (glutathione peroxidase-like enzyme I).
|
| |
Biochem J,
425,
513-522.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Spinks,
E.J.Shanks,
L.A.Cleghorn,
S.McElroy,
D.Jones,
D.James,
A.H.Fairlamb,
J.A.Frearson,
P.G.Wyatt,
and
I.H.Gilbert
(2009).
Investigation of trypanothione reductase as a drug target in Trypanosoma brucei.
|
| |
ChemMedChem,
4,
2060-2069.
|
 |
|
|
|
|
 |
G.A.Holloway,
W.N.Charman,
A.H.Fairlamb,
R.Brun,
M.Kaiser,
E.Kostewicz,
P.M.Novello,
J.P.Parisot,
J.Richardson,
I.P.Street,
K.G.Watson,
and
J.B.Baell
(2009).
Trypanothione reductase high-throughput screening campaign identifies novel classes of inhibitors with antiparasitic activity.
|
| |
Antimicrob Agents Chemother,
53,
2824-2833.
|
 |
|
|
|
|
 |
R.Perez-Pineiro,
A.Burgos,
D.C.Jones,
L.C.Andrew,
H.Rodriguez,
M.Suarez,
A.H.Fairlamb,
and
D.S.Wishart
(2009).
Development of a novel virtual screening cascade protocol to identify potential trypanothione reductase inhibitors.
|
| |
J Med Chem,
52,
1670-1680.
|
 |
|
|
|
|
 |
S.Patterson,
D.C.Jones,
E.J.Shanks,
J.A.Frearson,
I.H.Gilbert,
P.G.Wyatt,
and
A.H.Fairlamb
(2009).
Synthesis and evaluation of 1-(1-(Benzo[b]thiophen-2-yl)cyclohexyl)piperidine (BTCP) analogues as inhibitors of trypanothione reductase.
|
| |
ChemMedChem,
4,
1341-1353.
|
 |
|
|
|
|
 |
D.B.Castro-Pinto,
M.Genestra,
G.B.Menezes,
M.Waghabi,
A.Gonçalves,
C.De Nigris Del Cistia,
C.M.Sant'Anna,
L.L.Leon,
and
L.Mendonça-Lima
(2008).
Cloning and expression of trypanothione reductase from a New World Leishmania species.
|
| |
Arch Microbiol,
189,
375-384.
|
 |
|
|
|
|
 |
T.N.Gustafsson,
T.Sandalova,
J.Lu,
A.Holmgren,
and
G.Schneider
(2007).
High-resolution structures of oxidized and reduced thioredoxin reductase from Helicobacter pylori.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
833-843.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Jaeger,
and
L.Flohé
(2006).
The thiol-based redox networks of pathogens: unexploited targets in the search for new drugs.
|
| |
Biofactors,
27,
109-120.
|
 |
|
|
|
|
 |
E.I.Biterova,
A.A.Turanov,
V.N.Gladyshev,
and
J.J.Barycki
(2005).
Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism.
|
| |
Proc Natl Acad Sci U S A,
102,
15018-15023.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Hofmann,
H.J.Hecht,
and
L.Flohé
(2002).
Peroxiredoxins.
|
| |
Biol Chem,
383,
347-364.
|
 |
|
|
|
|
 |
P.A.van den Berg,
A.van Hoek,
C.D.Walentas,
R.N.Perham,
and
A.J.Visser
(1998).
Flavin fluorescence dynamics and photoinduced electron transfer in Escherichia coli glutathione reductase.
|
| |
Biophys J,
74,
2046-2058.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

