 |
PDBsum entry 1s5v

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.3.7
- 4-hydroxy-tetrahydrodipicolinate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate 4-semialdehyde + pyruvate = (2S,4S)-4-hydroxy-2,3,4,5- tetrahydrodipicolinate + H2O + H+
|
 |
 |
 |
 |
 |
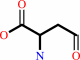 L-aspartate 4-semialdehyde
L-aspartate 4-semialdehyde
|
+
|
 pyruvate
pyruvate
|
=
|
(2S,4S)-4-hydroxy-2,3,4,5- tetrahydrodipicolinate
|
+
|
H2O
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
338:329-339
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of three site-directed mutants of Escherichia coli dihydrodipicolinate synthase: further evidence for a catalytic triad.
|
|
R.C.Dobson,
K.Valegård,
J.A.Gerrard.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Dihydrodipicolinate synthase (DHDPS, EC 4.2.1.52) catalyses the branchpoint
reaction of lysine biosynthesis in plants and microbes: the condensation of
(S)-aspartate-beta-semialdehyde and pyruvate. The crystal structure of wild-type
DHDPS has been published to 2.5A, revealing a tetrameric molecule comprised of
four identical (beta/alpha)(8)-barrels, each containing one active site.
Previous workers have hypothesised that the catalytic mechanism of the enzyme
involves a catalytic triad of amino acid residues, Tyr133, Thr44 and Tyr107,
which provide a proton shuttle to transport protons from the active site to
solvent. We have tested this hypothesis using site-directed mutagenesis to
produce three mutant enzymes: DHDPS-Y133F, DHDPS-T44V and DHDPS-Y107F. Each of
these mutants has substantially reduced activity, consistent with the catalytic
triad hypothesis. We have determined each mutant crystal structure to at least
2.35A resolution and compared the structures to the wild-type enzyme. All mutant
enzymes crystallised in the same space group as the wild-type form and only
minor differences in structure are observed. These results suggest that the
catalytic triad is indeed in operation in wild-type DHDPS.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Putative roles for the catalytic triad in the
mechanism of DHDPS. This mechanism has been adapted from Hutton
et al.[33.]
|
 |
Figure 5.
Figure 5. Stereo-view showing overlays of the active sites
of wild-type (black) and mutant DHDPS structures (gold).
Electron density covers the mutated residue in each image and is
contoured to 1s. Shown from top to bottom: a, DHDPS-Y133F; b,
DHDPS-T44V; c, DHDPS-Y107F. The images were generated using
O[31.] and Molray. [35.]
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2004,
338,
329-339)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Garg,
R.Tewari,
and
G.P.Raghava
(2010).
Virtual Screening of potential drug-like inhibitors against Lysine/DAP pathway of Mycobacterium tuberculosis.
|
| |
BMC Bioinformatics,
11,
S53.
|
 |
|
|
|
|
 |
J.E.Voss,
S.W.Scally,
N.L.Taylor,
S.C.Atkinson,
M.D.Griffin,
C.A.Hutton,
M.W.Parker,
M.R.Alderton,
J.A.Gerrard,
R.C.Dobson,
C.Dogovski,
and
M.A.Perugini
(2010).
Substrate-mediated stabilization of a tetrameric drug target reveals Achilles heel in anthrax.
|
| |
J Biol Chem,
285,
5188-5195.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.R.Devenish,
and
J.A.Gerrard
(2009).
The role of quaternary structure in (beta/alpha)(8)-barrel proteins: evolutionary happenstance or a higher level of structure-function relationships?
|
| |
Org Biomol Chem,
7,
833-839.
|
 |
|
|
|
|
 |
B.R.Burgess,
R.C.Dobson,
C.Dogovski,
G.B.Jameson,
M.W.Parker,
and
M.A.Perugini
(2008).
Purification, crystallization and preliminary X-ray diffraction studies to near-atomic resolution of dihydrodipicolinate synthase from methicillin-resistant Staphylococcus aureus.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
659-661.
|
 |
|
|
|
|
 |
B.R.Burgess,
R.C.Dobson,
M.F.Bailey,
S.C.Atkinson,
M.D.Griffin,
G.B.Jameson,
M.W.Parker,
J.A.Gerrard,
and
M.A.Perugini
(2008).
Structure and evolution of a novel dimeric enzyme from a clinically important bacterial pathogen.
|
| |
J Biol Chem,
283,
27598-27603.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.P.Phenix,
K.Nienaber,
P.H.Tam,
L.T.Delbaere,
and
D.R.Palmer
(2008).
Structural, functional and calorimetric investigation of MosA, a dihydrodipicolinate synthase from Sinorhizobium meliloti l5-30, does not support involvement in rhizopine biosynthesis.
|
| |
Chembiochem,
9,
1591-1602.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.C.Dobson,
M.D.Griffin,
S.R.Devenish,
F.G.Pearce,
C.A.Hutton,
J.A.Gerrard,
G.B.Jameson,
and
M.A.Perugini
(2008).
Conserved main-chain peptide distortions: a proposed role for Ile203 in catalysis by dihydrodipicolinate synthase.
|
| |
Protein Sci,
17,
2080-2090.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.C.Dobson,
S.C.Atkinson,
M.A.Gorman,
J.M.Newman,
M.W.Parker,
and
M.A.Perugini
(2008).
The purification, crystallization and preliminary X-ray diffraction analysis of dihydrodipicolinate synthase from Clostridium botulinum.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
206-208.
|
 |
|
|
|
|
 |
S.R.Devenish,
J.A.Gerrard,
G.B.Jameson,
and
R.C.Dobson
(2008).
The high-resolution structure of dihydrodipicolinate synthase from Escherichia coli bound to its first substrate, pyruvate.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
1092-1095.
|
 |
|
|
|
|
 |
S.Wolterink-van Loo,
M.Levisson,
M.C.Cabrières,
M.C.Franssen,
and
J.van der Oost
(2008).
Characterization of a thermostable dihydrodipicolinate synthase from Thermoanaerobacter tengcongensis.
|
| |
Extremophiles,
12,
461-469.
|
 |
|
|
|
|
 |
C.A.Hutton,
M.A.Perugini,
and
J.A.Gerrard
(2007).
Inhibition of lysine biosynthesis: an evolving antibiotic strategy.
|
| |
Mol Biosyst,
3,
458-465.
|
 |
|
|
|
|
 |
E.Blagova,
V.Levdikov,
N.Milioti,
M.J.Fogg,
A.K.Kalliomaa,
J.A.Brannigan,
K.S.Wilson,
and
A.J.Wilkinson
(2006).
Crystal structure of dihydrodipicolinate synthase (BA3935) from Bacillus anthracis at 1.94 A resolution.
|
| |
Proteins,
62,
297-301.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.A.Azevedo,
M.Lancien,
and
P.J.Lea
(2006).
The aspartic acid metabolic pathway, an exciting and essential pathway in plants.
|
| |
Amino Acids,
30,
143-162.
|
 |
|
|
|
|
 |
S.Watanabe,
N.Shimada,
K.Tajima,
T.Kodaki,
and
K.Makino
(2006).
Identification and characterization of L-arabonate dehydratase, L-2-keto-3-deoxyarabonate dehydratase, and L-arabinolactonase involved in an alternative pathway of L-arabinose metabolism. Novel evolutionary insight into sugar metabolism.
|
| |
J Biol Chem,
281,
33521-33536.
|
 |
|
|
|
|
 |
M.A.Perugini,
M.D.Griffin,
B.J.Smith,
L.E.Webb,
A.J.Davis,
E.Handman,
and
J.A.Gerrard
(2005).
Insight into the self-association of key enzymes from pathogenic species.
|
| |
Eur Biophys J,
34,
469-476.
|
 |
|
|
|
|
 |
R.C.Dobson,
M.D.Griffin,
G.B.Jameson,
and
J.A.Gerrard
(2005).
The crystal structures of native and (S)-lysine-bound dihydrodipicolinate synthase from Escherichia coli with improved resolution show new features of biological significance.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1116-1124.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

