 |
PDBsum entry 1l1q

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.7
- adenine phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Ribose activation
|
 |
 |
 |
 |
 |
Reaction:
|
 |
AMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + adenine
|
 |
 |
 |
 |
 |
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
=
|
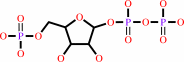 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
+
|
adenine
Bound ligand (Het Group name = )
matches with 81.82% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:39981-39988
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Closed site complexes of adenine phosphoribosyltransferase from Giardia lamblia reveal a mechanism of ribosyl migration.
|
|
W.Shi,
A.E.Sarver,
C.C.Wang,
K.S.Tanaka,
S.C.Almo,
V.L.Schramm.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The adenine phosphoribosyltransferase (APRTase) from Giardia lamblia was
co-crystallized with 9-deazaadenine and sulfate or with 9-deazaadenine and
Mg-phosphoribosylpyrophosphate. The complexes were solved and refined to 1.85
and 1.95 A resolution. Giardia APRTase is a symmetric homodimer with the
monomers built around Rossman fold cores, an element common to all known purine
phosphoribosyltransferases. The catalytic sites are capped with a small hood
domain that is unique to the APRTases. These structures reveal several features
relevant to the catalytic function of APRTase: 1) a non-proline cis peptide bond
(Glu(61)-Ser(62)) is required to form the pyrophosphate binding site in the
APRTase.9dA.MgPRPP complex but is a trans peptide bond in the absence of
pyrophosphate group, as observed in the APRTase.9dA.SO4 complex; 2) a catalytic
site loop is closed and fully ordered in both complexes, with Glu(100) from the
catalytic loop acting as the acid/base for protonation/deprotonation of N-7 of
the adenine ring; 3) the pyrophosphoryl charge is neutralized by a single Mg2+
ion and Arg(63), in contrast to the hypoxanthine-guanine
phosphoribosyltransferases, which use two Mg2+ ions; and 4) the nearest
structural neighbors to APRTases are the orotate phosphoribosyltransferases,
suggesting different paths of evolution for adenine relative to other purine
PRTases. An overlap comparison of AMP and 9-deazaadenine plus Mg-PRPP at the
catalytic sites of APRTases indicated that reaction coordinate motion involves a
2.1-A excursion of the ribosyl anomeric carbon, whereas the adenine ring and the
5-phosphoryl group remained fixed. G. lamblia APRTase therefore provides another
example of nucleophilic displacement by electrophile migration.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Fig. 5. Peptide conformation of Glu61-Ser62. a,
stereoview of trans conformation of Glu61-Ser62 in the Giardia
APRTase·9dA·SO[4] complex. The peptide segment
connecting Glu61 and Ser62 is stabilized in the trans
conformation by a sulfate ion. b, stereoview of the cis
conformation of the same peptide in
APRTase·9dA·MgPRPP complex. The unusual cis
conformation in the peptide link between Glu61 and Ser62 orients
the amide nitrogen atoms of Ser62 and Arg63 to bind PRPP.
|
 |
Figure 7.
Fig. 7. Hydrogen bond network in Giardia
APRTase·9dA·MgPRPP complex. Distances (<3.2
Å) are in angstroms.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
39981-39988)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.R.Zomorrodi,
and
C.D.Maranas
(2010).
Improving the iMM904 S. cerevisiae metabolic model using essentiality and synthetic lethality data.
|
| |
BMC Syst Biol,
4,
178.
|
 |
|
|
|
|
 |
R.Takahashi,
S.Nakamura,
T.Nakazawa,
K.Minoura,
T.Yoshida,
Y.Nishi,
Y.Kobayashi,
and
T.Ohkubo
(2010).
Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase.
|
| |
J Biochem,
147,
95.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.S.Burgos,
M.C.Ho,
S.C.Almo,
and
V.L.Schramm
(2009).
A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT.
|
| |
Proc Natl Acad Sci U S A,
106,
13748-13753.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.H.Rehse,
and
T.H.Tahirov
(2005).
Crystal structure of a purine/pyrimidine phosphoribosyltransferase-related protein from Thermus thermophilus HB8.
|
| |
Proteins,
61,
658-665.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.H.el Kouni
(2003).
Potential chemotherapeutic targets in the purine metabolism of parasites.
|
| |
Pharmacol Ther,
99,
283-309.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

