|
Figure 2.
Figure 2 Predicted folding of a generic 5' hammerhead
ribozyme-SRP RNA-3' hepatitis 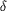 virus
ribozyme transcript (in this case, the sequence of the 45 RNA
construct is shown in red). Bases of the transcription plasmid
insert important for efficient T7 RNA polymerase transcription
and ribozyme activity are boxed in white and black,
respectively. Cloning sites are in brackets and 5' and 3'
ribozyme cleavage sites are indicated by black arrows.
Constructs of this type were cloned into the XbaI/PstI sites of
a pUC18-based vector carrying a 5' T7 promoter and a 3' fragment
of an hepatitis virus
ribozyme transcript (in this case, the sequence of the 45 RNA
construct is shown in red). Bases of the transcription plasmid
insert important for efficient T7 RNA polymerase transcription
and ribozyme activity are boxed in white and black,
respectively. Cloning sites are in brackets and 5' and 3'
ribozyme cleavage sites are indicated by black arrows.
Constructs of this type were cloned into the XbaI/PstI sites of
a pUC18-based vector carrying a 5' T7 promoter and a 3' fragment
of an hepatitis 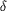 virus
ribozyme sequence (pUC18T7Pst virus
ribozyme sequence (pUC18T7Pst 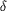 V;
Dr Sandra Searles, MRC-LMB, personal communication). This
created a new 5' hammerhead ribozyme sequence and reconstituted
a complete 3' hepatitis V;
Dr Sandra Searles, MRC-LMB, personal communication). This
created a new 5' hammerhead ribozyme sequence and reconstituted
a complete 3' hepatitis 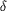 virus
ribozyme sequence. After digestion of the resulting plasmid at
the 3' HindIII site (indicated by a blue arrow) and in vitro
run-off transcription/co-transcriptional RNA 5'-end cleavage by
hammerhead ribozyme (Price, Oubridge et al., 1998[Price, S. R.,
Oubridge, C., Varani, G. & Nagai, K. (1998). RNA-Protein
Interactions: A Practical Approach, edited by C. Smith, pp.
37-74. Oxford University Press.]), 3'-end cleavage of the RNA
product by the hepatitis virus
ribozyme sequence. After digestion of the resulting plasmid at
the 3' HindIII site (indicated by a blue arrow) and in vitro
run-off transcription/co-transcriptional RNA 5'-end cleavage by
hammerhead ribozyme (Price, Oubridge et al., 1998[Price, S. R.,
Oubridge, C., Varani, G. & Nagai, K. (1998). RNA-Protein
Interactions: A Practical Approach, edited by C. Smith, pp.
37-74. Oxford University Press.]), 3'-end cleavage of the RNA
product by the hepatitis 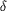 virus
ribozyme required a 2 min annealing step at 338 K followed by
a 4 h incubation at 328 K. virus
ribozyme required a 2 min annealing step at 338 K followed by
a 4 h incubation at 328 K.
|
