 |
PDBsum entry 9hg0

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
9hg0
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of m. Smegmatis gmp reductase with xmp Intermidiate in complex with NADP+ and imp.
|
|
Structure:
|
 |
Gmp reductase. Chain: a, b, c, d, e, f, g, h. Synonym: guanosine 5'-monophosphate reductase,gmpr. Engineered: yes
|
|
Source:
|
 |
Mycolicibacterium smegmatis. Organism_taxid: 1772. Strain: atcc 700084 / mc(2)155. Gene: guab1, msmeg_3634, msmei_3548. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.220
|
R-free:
|
0.249
|
|
|
Authors:
|
 |
M.Dolezal,I.Pichova
|
|
Key ref:
|
 |
M.Dolezal
et al.
Structural basis for allosteric regulation of mycobac guanosine 5'-Monophosphate reductase with ATP and gtp.
To be published,
.
|
 |
|
Date:
|
 |
|
18-Nov-24
|
Release date:
|
27-Aug-25
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A0QYE8
(A0QYE8_MYCS2) -
GMP reductase from Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
478 a.a.
473 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.7.1.7
- Gmp reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + NH4+ + NADP+ = GMP + NADPH + 2 H+
|
 |
 |
 |
 |
 |
IMP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NH4(+)
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
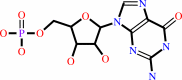 GMP
GMP
|
+
|
 NADPH
NADPH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

