 |
PDBsum entry 9gv8
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
N-acyl-d-amino-acid deacylase (d-acylase) from klebsiella pneumoniae in the absence of glycerol
|
|
Structure:
|
 |
Amidohydrolase family protein. Chain: a. Synonym: d-aminoacylase. Engineered: yes
|
|
Source:
|
 |
Klebsiella pneumoniae subsp. Pneumoniae kp13. Organism_taxid: 1123862. Gene: dan, b5l96_17380, b6i68_16395, banra_04485, bl124_00028775, dw281_07100, eao17_17410, fxn67_23300, g4v31_23420, gjj08_023370, gjj13_021835, gjj18_03715, gnf00_21165, nctc11679_04932, nctc13443_01470, nctc5052_02702, nctc9601_05682, nctc9661_05441, qig75_04865, samea3499874_02915, samea3499901_00964, samea3512100_03687, samea3515122_03158, samea3538658_01149, samea3538828_01003, samea3673026_03520, samea3720909_04521,
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.228
|
R-free:
|
0.269
|
|
|
Authors:
|
 |
J.A.Gavira,S.Martinez-Rodriguez
|
|
Key ref:
|
 |
S.Martinez-Rodriguez
and
j.a.gavira
(2025).
Revisiting d-Acylases for d-Amino acid production..
Microb biotechnol,
18,
70179.
PubMed id:
|
 |
|
Date:
|
 |
|
23-Sep-24
|
Release date:
|
18-Jun-25
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
W9BIW9
(W9BIW9_KLEPN) -
Amidohydrolase family protein from Klebsiella pneumoniae
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
479 a.a.
478 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.5.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.5.1.81
- N-acyl-D-amino-acid deacylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-acyl-D-amino acid + H2O = a D-alpha-amino acid + a carboxylate
|
 |
 |
 |
 |
 |
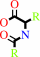 N-acyl-D-amino acid
N-acyl-D-amino acid
|
+
|
H2O
|
=
|
D-alpha-amino acid
|
+
|
 carboxylate
carboxylate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

