 |
PDBsum entry 9grh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Biosynthetic protein
|
PDB id
|
|

|

|
9grh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.4.24
- diphosphoinositol-pentakisphosphate 1-kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
1D-myo-inositol hexakisphosphate + ATP = 1-diphospho-1D-myo-inositol 2,3,4,5,6-pentakisphosphate + ADP
|
|
2.
|
5-diphospho-1D-myo-inositol 1,2,3,4,6-pentakisphosphate + ATP + H+ = 1,5-bis(diphospho)-1D-myo-inositol 2,3,4,6-tetrakisphosphate + ADP
|
|
 |
 |
 |
 |
 |
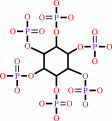 1D-myo-inositol hexakisphosphate
1D-myo-inositol hexakisphosphate
|
+
|
 ATP
ATP
|
=
|
1-diphospho-1D-myo-inositol 2,3,4,5,6-pentakisphosphate
|
+
|
 ADP
ADP
|
|
 |
 |
 |
 |
 |
5-diphospho-1D-myo-inositol 1,2,3,4,6-pentakisphosphate
|
+
|
 ATP
ATP
|
+
|
H(+)
|
=
|
1,5-bis(diphospho)-1D-myo-inositol 2,3,4,6-tetrakisphosphate
|
+
|
 ADP
ADP
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

