 |
PDBsum entry 9byc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
9byc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Class 12 model for product condition of bacillus subtilis ribonucleotide reductase complex
|
|
Structure:
|
 |
Ribonucleoside-diphosphate reductase subunit alpha. Chain: a, b. Synonym: ribonucleotide reductase large subunit. Engineered: yes. Ribonucleoside-diphosphate reductase subunit beta. Chain: c, d. Synonym: ribonucleotide reductase small subunit. Engineered: yes
|
|
Source:
|
 |
Bacillus subtilis. Organism_taxid: 1423. Gene: nrde, nrda, bsu17380. Expressed in: escherichia coli. Expression_system_taxid: 562. Gene: nrdf, bsu17390. Expression_system_taxid: 562
|
|
Authors:
|
 |
D.Xu,W.C.Thomas,A.A.Burnim,N.Ando
|
|
Key ref:
|
 |
D.Xu
et al.
(2025).
Conformational landscapes of a class i ribonucleotide reductase complex during turnover reveal intrinsic dy and asymmetry.
Nat commun,
16,
2458.
PubMed id:
|
 |
|
Date:
|
 |
|
23-May-24
|
Release date:
|
19-Mar-25
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D:
E.C.1.17.4.1
- ribonucleoside-diphosphate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2'-deoxyribonucleoside 5'-diphosphate + [thioredoxin]-disulfide + H2O = a ribonucleoside 5'-diphosphate + [thioredoxin]-dithiol
|
 |
 |
 |
 |
 |
 2'-deoxyribonucleoside diphosphate
2'-deoxyribonucleoside diphosphate
|
+
|
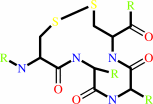 thioredoxin disulfide
thioredoxin disulfide
|
+
|
H(2)O
|
=
|
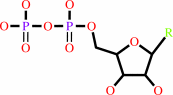 ribonucleoside diphosphate
ribonucleoside diphosphate
|
+
|
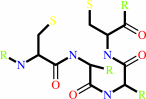 thioredoxin
thioredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(3+) or adenosylcob(III)alamin or Mn(2+)
|
 |
 |
 |
 |
 |
Fe(3+)
|
or
|
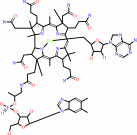 adenosylcob(III)alamin
adenosylcob(III)alamin
|
or
|
Mn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

