 |
PDBsum entry 9b9m

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
9b9m
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of iron-bound flcd from pseudomonas aeruginosa
|
|
Structure:
|
 |
Pyrroloquinoline quinone (coenzyme pqq) biosynthesis protein c. Chain: a, b, c, d. Synonym: pyrroloquinoline-quinone synthase. Engineered: yes
|
|
Source:
|
 |
Pseudomonas aeruginosa. Organism_taxid: 287. Gene: pqqc_2, caz10_19980, dt376_18990, nctc13621_03444, paerug_p19_london_7_vim_2_05_10_05324. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.07Å
|
R-factor:
|
0.200
|
R-free:
|
0.241
|
|
|
Authors:
|
 |
M.E.Walker,T.L.Grove,B.Li,M.R.Redinbo
|
|
Key ref:
|
 |
W.C.Simke
et al.
Structural basis for methine excision by a heme oxygenase-Like enzyme.
Acs cent sci,
.
PubMed id:
|
 |
|
Date:
|
 |
|
02-Apr-24
|
Release date:
|
31-Jul-24
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A0A0C7AN42
(A0A0C7AN42_PSEAI) -
Pyrroloquinoline quinone (Coenzyme PQQ) biosynthesis protein C from Pseudomonas aeruginosa
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
328 a.a.
326 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.3.11
- pyrroloquinoline-quinone synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
6-(2-amino-2-carboxyethyl)-7,8-dioxo-1,2,3,4,7,8-hexahydroquinoline-2,4- dicarboxylate + 3 O2 = pyrroloquinoline quinone + 2 H2O2 + 2 H2O + H+
|
 |
 |
 |
 |
 |
6-(2-amino-2-carboxyethyl)-7,8-dioxo-1,2,3,4,7,8-hexahydroquinoline-2,4- dicarboxylate
|
+
|
 3
×
O2
3
×
O2
|
=
|
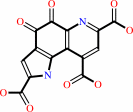 pyrroloquinoline quinone
pyrroloquinoline quinone
|
+
|
 2
×
H2O2
2
×
H2O2
|
+
|
2
×
H2O
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

