 |
PDBsum entry 8vmc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Biosynthetic protein
|
PDB id
|
|

|

|
8vmc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Biosynthetic protein
|
 |
|
Title:
|
 |
Composite structure of human fasn with NADPH in state 6
|
|
Structure:
|
 |
Fatty acid synthase. Chain: a, b. Synonym: type i fatty acid synthase. Ec: 2.3.1.85,2.3.1.38,2.3.1.39,2.3.1.41,1.1.1.100,4.2.1.59,1.3.1.39, 3.1.2.14. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: fasn, fas. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108. Expression_system_cell_line: sf9
|
|
Authors:
|
 |
K.Schultz,R.Marmorstein
|
|
Key ref:
|
 |
K.Schultz
et al.
Snapshots of acyl carrier protein shuttling in human acid synthase..
Nature,
.
PubMed id:
|
 |
|
Date:
|
 |
|
13-Jan-24
|
Release date:
|
26-Feb-25
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P49327
(FAS_HUMAN) -
Fatty acid synthase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
2511 a.a.
2068 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.100
- 3-oxoacyl-[acyl-carrier-protein] reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a (3R)-hydroxyacyl-[ACP] + NADP+ = a 3-oxoacyl-[ACP] + NADPH + H+
|
 |
 |
 |
 |
 |
(3R)-hydroxyacyl-[ACP]
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
3-oxoacyl-[ACP]
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.3.1.39
- enoyl-[acyl-carrier-protein] reductase (Nadph, Re-specific).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2,3-saturated acyl-[ACP] + NADP+ = a (2E)-enoyl-[ACP] + NADPH + H+
|
 |
 |
 |
 |
 |
2,3-saturated acyl-[ACP]
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
(2E)-enoyl-[ACP]
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.3.1.38
- [acyl-carrier-protein] S-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
holo-[ACP] + acetyl-CoA = acetyl-[ACP] + CoA
|
 |
 |
 |
 |
 |
holo-[ACP]
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
acetyl-[ACP]
|
+
|
CoA
Bound ligand (Het Group name = )
matches with 52.38% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.2.3.1.39
- [acyl-carrier-protein] S-malonyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
holo-[ACP] + malonyl-CoA = malonyl-[ACP] + CoA
|
 |
 |
 |
 |
 |
holo-[ACP]
|
+
|
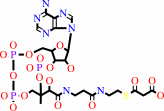 malonyl-CoA
malonyl-CoA
|
=
|
malonyl-[ACP]
|
+
|
CoA
Bound ligand (Het Group name = )
matches with 52.38% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.2.3.1.41
- beta-ketoacyl-[acyl-carrier-protein] synthase I.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a fatty acyl-[ACP] + malonyl-[ACP] + H+ = a 3-oxoacyl-[ACP] + holo- [ACP] + CO2
|
 |
 |
 |
 |
 |
fatty acyl-[ACP]
|
+
|
malonyl-[ACP]
|
+
|
H(+)
|
=
|
3-oxoacyl-[ACP]
|
+
|
holo- [ACP]
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
E.C.2.3.1.85
- fatty-acid synthase system.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
acetyl-CoA + n malonyl-CoA + 2n NADPH + 2n H+ = a long-chain fatty acid + (n+1) CoA + n CO2 + 2n NADP+
|
 |
 |
 |
 |
 |
acetyl-CoA
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
 n
malonyl-CoA
n
malonyl-CoA
|
+
|
2n NADPH
|
+
|
2n H(+)
|
=
|
 long-chain fatty acid
long-chain fatty acid
|
+
|
(n+1) CoA
|
+
|
 n
CO2
n
CO2
|
+
|
2n NADP(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 7:
|
 |
E.C.3.1.2.14
- oleoyl-[acyl-carrier-protein] hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(9Z)-octadecenoyl-[ACP] + H2O = (9Z)-octadecenoate + holo-[ACP] + H+
|
 |
 |
 |
 |
 |
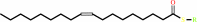 Oleoyl-[acyl-carrier-protein]
Oleoyl-[acyl-carrier-protein]
|
+
|
n
H(2)O
|
=
|
 [acyl-carrier-protein]
[acyl-carrier-protein]
|
+
|
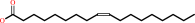 oleate
oleate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 8:
|
 |
E.C.4.2.1.59
- 3-hydroxyacyl-[acyl-carrier-protein] dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a (3R)-hydroxyacyl-[ACP] = a (2E)-enoyl-[ACP] + H2O
|
 |
 |
 |
 |
 |
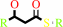 (3R)-3-hydroxyacyl-[acyl-carrier protein]
(3R)-3-hydroxyacyl-[acyl-carrier protein]
|
=
|
n
trans-2-enoyl-[acyl- carrier protein]
|
+
|
H(2)O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

