 |
PDBsum entry 8spk
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of antarctic pet-degrading enzyme
|
|
Structure:
|
 |
Lipase 1. Chain: a, b. Synonym: triacylglycerol lipase. Engineered: yes
|
|
Source:
|
 |
Moraxella. Organism_taxid: 475. Gene: lip1, l1. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.60Å
|
R-factor:
|
0.154
|
R-free:
|
0.180
|
|
|
Authors:
|
 |
A.A.Furtado,P.Blazquez-Sanchez,A.Grinen,J.A.Vargas,D.A.Leonardo, S.A.Sculaccio,H.M.Pereira,B.Diez,R.C.Garratt,C.A.Ramirez-Sarmiento
|
|
Key ref:
|
 |
P.Blazquez-Sanchez
et al.
Engineering the catalytic activity of an antarctic pet-Degrading enzyme by loop exchange..
Protein sci,
.
PubMed id:
|
 |
|
Date:
|
 |
|
03-May-23
|
Release date:
|
23-Aug-23
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P19833
(LIP1_MORS1) -
Lipase 1 from Moraxella sp. (strain TA144)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
319 a.a.
265 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.1.3
- triacylglycerol lipase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a triacylglycerol + H2O = a diacylglycerol + a fatty acid + H+
|
 |
 |
 |
 |
 |
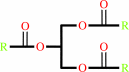 triacylglycerol
triacylglycerol
|
+
|
H2O
|
=
|
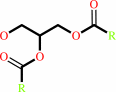 diacylglycerol
diacylglycerol
|
+
|
 fatty acid
fatty acid
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

