 |
PDBsum entry 8sml

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/immune system
|
PDB id
|
|

|

|
8sml
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 570 a.a.
570 a.a.
|
 |

|

|

|

|

|

|

|

 106 a.a.
106 a.a.
|
 |

|

|

|

|

|

|

|

 121 a.a.
121 a.a.
|
 |

|

|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase/immune system
|
 |
|
Title:
|
 |
Hpad4 bound to inhibitory fab hi365
|
|
Structure:
|
 |
Protein-arginine deiminase type-4. Chain: a, d. Synonym: hl-60 pad,peptidylarginine deiminase iv,protein-arginine deiminase type iv. Engineered: yes. Fab hi365 light chain. Chain: b, e. Engineered: yes. Fab hi365 heavy chain.
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Variant: 9606. Gene: padi4, pad4, padi5, pdi5. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_atcc_number: 562. Expression_system_taxid: 562
|
|
Authors:
|
 |
A.Maker,K.A.Verba
|
|
Key ref:
|
 |
X.Zhou
et al.
Antibody discovery identifies regulatory mechanisms o protein arginine deiminase 4..
Nat chem biol,
.
PubMed id:
|
 |
|
Date:
|
 |
|
26-Apr-23
|
Release date:
|
06-Mar-24
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9UM07
(PADI4_HUMAN) -
Protein-arginine deiminase type-4 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
663 a.a.
570 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, D:
E.C.3.5.3.15
- protein-arginine deiminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-arginyl-[protein] + H2O = L-citrullyl-[protein] + NH4+
|
 |
 |
 |
 |
 |
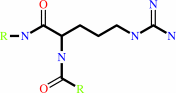 Protein L-arginine
Protein L-arginine
|
+
|
H(2)O
|
=
|
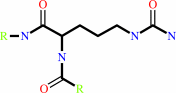 protein L-citrulline
protein L-citrulline
|
+
|
NH(3)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
| |

