 |
PDBsum entry 8sfa
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of the engineered ssopox variant iiic1
|
|
Structure:
|
 |
Aryldialkylphosphatase. Chain: d, s, b, t. Synonym: paraoxonase,ssopox,phosphotriesterase-like lactonase. Engineered: yes
|
|
Source:
|
 |
Saccharolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2). Organism_taxid: 273057. Strain: atcc 35092 / dsm 1617 / jcm 11322 / p2. Variant: iiic1. Gene: php, sso2522. Expressed in: escherichia coli 'bl21-gold(de3)plyss ag'. Expression_system_taxid: 866768
|
|
Resolution:
|
 |
|
2.32Å
|
R-factor:
|
0.193
|
R-free:
|
0.242
|
|
|
Authors:
|
 |
P.Jacquet,R.Billot,A.Shimon,N.Hoekstra,C.Bergonzi,A.Jenks,D.Daude, M.H.Elias
|
|
Key ref:
|
 |
P.Jacquet
et al.
Changes in active site loops conformation relates to transition from lactonase to phosphotriesterase.
To be published,
.
PubMed id:
|
 |
|
Date:
|
 |
|
10-Apr-23
|
Release date:
|
17-Apr-24
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q97VT7
(PHP_SULSO) -
Aryldialkylphosphatase from Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
314 a.a.
314 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 6 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.8.1
- aryldialkylphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
An aryl dialkyl phosphate + H2O = dialkyl phosphate + an aryl alcohol
|
 |
 |
 |
 |
 |
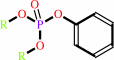 aryl dialkyl phosphate
aryl dialkyl phosphate
|
+
|
H2O
|
=
|
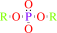 dialkyl phosphate
dialkyl phosphate
|
+
|
aryl alcohol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

