 |
PDBsum entry 8oyc
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Time-resolved sfx structure of the class ii photolyase complexed with a thymine dimer (100 microsecond timpeoint)
|
|
Structure:
|
 |
Deoxyribodipyrimidine photo-lyase. Chain: a, b. Synonym: DNA photolyase. Engineered: yes. Cpd-comprising oligonucleotide. Chain: c, e. Engineered: yes. Counterstrand-oligonucleotide. Chain: d, f.
|
|
Source:
|
 |
Methanosarcina mazei go1. Organism_taxid: 192952. Gene: mm_0852. Expressed in: methanosarcina mazei go1. Expression_system_taxid: 192952. Synthetic: yes. Synthetic construct. Organism_taxid: 32630. Organism_taxid: 32630
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.307
|
R-free:
|
0.371
|
|
|
Authors:
|
 |
T.J.Lane,N.-E.Christou,D.V.M.Melo,V.Apostolopoulou,A.Pateras, A.R.Mashhour,M.Galchenkova,S.Gunther,P.Reinke,V.Kremling, D.Oberthuer,A.Henkel,J.Sprenger,T.E.S.Scheer,E.Lange,O.N.Yefanov, P.Middendorf,J.A.Sellberg,R.Schubert,A.Fadini,C.Cirelli,E.V.Beale, P.Johnson,F.Dworkowski,D.Ozerov,Q.Bertrand,M.Wranik,E.D.Zitter, D.Turk,S.Bajt,H.Chapman,C.Bacellar
|
|
Key ref:
|
 |
T.J.Lane
et al.
Time-Resolved crystallography captures light-Driven d repair.
Science,
.
PubMed id:
|
 |
|
Date:
|
 |
|
03-May-23
|
Release date:
|
22-Nov-23
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.4.1.99.3
- deoxyribodipyrimidine photo-lyase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
EC 4.1.99.3
|
 |
 |
 |
 |
 |
Reaction:
|
 |
cyclobutadipyrimidine (in DNA) = 2 pyrimidine residues (in DNA)
|
 |
 |
 |
 |
 |
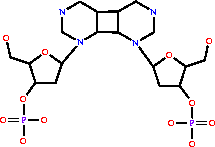 cyclobutadipyrimidine (in DNA)
cyclobutadipyrimidine (in DNA)
|
=
|
2
×
pyrimidine residues (in DNA)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
5,10-methenyltetrahydrofolate; FAD
|
 |
 |
 |
 |
 |
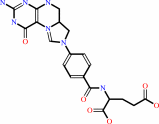 5,10-methenyltetrahydrofolate
5,10-methenyltetrahydrofolate
|
FAD
Bound ligand (Het Group name =
FDA)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

