 |
PDBsum entry 8hfd
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of allantoinase from e. Coli bl21
|
|
Structure:
|
 |
Allantoinase. Chain: a, b, c, d. Synonym: allantoin-utilizing enzyme. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Gene: allb, bmt91_19075, e2127_17875, e2128_18450, e2132_17365, e4t14_14655, e6d34_22765, elt21_17110. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008
|
|
Resolution:
|
 |
|
2.07Å
|
R-factor:
|
0.206
|
R-free:
|
0.249
|
|
|
Authors:
|
 |
E.S.Lin,H.Y.Huang,P.C.Yang,H.W.Liu,C.Y.Huang
|
|
Key ref:
|
 |
Y.H.Huang
et al.
(2023).
Crystal structure of allantoinase from escherichia co bl21: a molecular insight into a role of the active s loops in catalysis..
Molecules,
28,
.
PubMed id:
|
 |
|
Date:
|
 |
|
10-Nov-22
|
Release date:
|
18-Oct-23
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P77671
(ALLB_ECOLI) -
Allantoinase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
453 a.a.
452 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.2.5
- allantoinase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP Catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-allantoin + H2O = allantoate + H+
|
 |
 |
 |
 |
 |
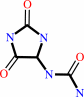 (S)-allantoin
(S)-allantoin
|
+
|
H2O
|
=
|
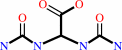 allantoate
allantoate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

