 |
PDBsum entry 8hfc
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Cryo-em structure of yeast erf2/erf4 complex
|
|
Structure:
|
 |
Palmitoyltransferase erf2. Chain: a. Synonym: dhhc cysteine-rich domain-containing protein erf2,ras protein acyltransferase. Engineered: yes. Ras modification protein erf4. Chain: b. Synonym: hyperactive ras suppressor protein 5. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae (strain atcc 204508 / s288c). Baker's yeast. Organism_taxid: 559292. Gene: erf2, ylr246w. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108. Gene: shr5, erf4, yol110w, hrc237. Expression_system_taxid: 7108
|
|
Authors:
|
 |
J.Wu,Q.Hu,Y.Zhang,A.Yang,S.Liu
|
|
Key ref:
|
 |
J.Wu
et al.
Cryo-Em structure of yeast erf2/erf4 complex.
To be published,
.
PubMed id:
|
 |
|
Date:
|
 |
|
10-Nov-22
|
Release date:
|
22-Nov-23
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain A:
E.C.2.3.1.225
- protein S-acyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-cysteinyl-[protein] + hexadecanoyl-CoA = S-hexadecanoyl-L-cysteinyl- [protein] + CoA
|
 |
 |
 |
 |
 |
L-cysteinyl-[protein]
|
+
|
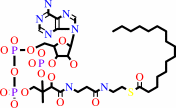 hexadecanoyl-CoA
hexadecanoyl-CoA
|
=
|
S-hexadecanoyl-L-cysteinyl- [protein]
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

