 |
PDBsum entry 8hbe

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Signaling protein
|
PDB id
|
|

|

|
8hbe
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Signaling protein
|
 |
|
Title:
|
 |
Structure of human soluble guanylate cyclase in the inactive state at 3.1 angstrom
|
|
Structure:
|
 |
Guanylate cyclase soluble subunit alpha-1. Chain: a. Synonym: gcs-alpha-1,guanylate cyclase soluble subunit alpha-3,gcs- alpha-3,soluble guanylate cyclase large subunit. Engineered: yes. Other_details: genbank: aah28384.1. Guanylate cyclase soluble subunit beta-1. Chain: b. Synonym: gcs-beta-1,guanylate cyclase soluble subunit beta-3,gcs-
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: gucy1a1, guc1a3, gucsa3, gucy1a3. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108. Gene: gucy1b1, guc1b3, gucsb3, gucy1b3. Expression_system_taxid: 7108
|
|
Authors:
|
 |
L.Chen,R.Liu
|
|
Key ref:
|
 |
R.Liu
et al.
(2023).
No binds to the distal site of haem in the fully acti soluble guanylate cyclase..
Nitric oxide,
135,
17.
PubMed id:
|
 |
|
Date:
|
 |
|
28-Oct-22
|
Release date:
|
19-Apr-23
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.4.6.1.2
- guanylate cyclase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
GTP = 3',5'-cyclic GMP + diphosphate
|
 |
 |
 |
 |
 |
 GTP
GTP
|
=
|
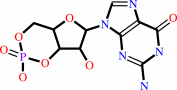 3',5'-cyclic GMP
3',5'-cyclic GMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

