 |
PDBsum entry 8guh
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Serine palmitoyltransferase from sphingobacterium multivorum complexed with tris
|
|
Structure:
|
 |
Serine palmitoyltransferase. Chain: a. Synonym: spt. Engineered: yes
|
|
Source:
|
 |
Sphingobacterium multivorum. Organism_taxid: 28454. Gene: spt, i6j33_20140, nctc11343_02561, sphingo8bc_150128. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.65Å
|
R-factor:
|
0.170
|
R-free:
|
0.214
|
|
|
Authors:
|
 |
T.Murakami,A.Takahashi,A.Katayama,I.Miyahara,N.Kamiya,H.Ikushiro, T.Yano
|
|
Key ref:
|
 |
H.Ikushiro
et al.
(2022).
Crystal structure of sphingobacterium multivorum seri palmitoyltransferase complexed with tris(hydroxymethyl)aminomethane..
Acta crystallogr ,Sect f,
78,
408.
PubMed id:
|
 |
|
Date:
|
 |
|
12-Sep-22
|
Release date:
|
19-Jul-23
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A7BFV6
(SPT_SPHMU) -
Serine palmitoyltransferase from Sphingobacterium multivorum
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
399 a.a.
394 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.50
- serine C-palmitoyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-serine + hexadecanoyl-CoA + H+ = 3-oxosphinganine + CO2 + CoA
|
 |
 |
 |
 |
 |
 L-serine
L-serine
|
+
|
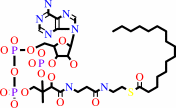 hexadecanoyl-CoA
hexadecanoyl-CoA
|
+
|
H(+)
|
=
|
3-oxosphinganine
|
+
|
CO2
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
K9E)
matches with 62.50% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

