 |
PDBsum entry 8gss

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.18
- glutathione transferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
RX + glutathione = an S-substituted glutathione + a halide anion + H+
|
 |
 |
 |
 |
 |
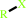 RX
RX
|
+
|
glutathione
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
S-substituted glutathione
|
+
|
halide anion
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
274:84
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
|
|
A.J.Oakley,
M.Lo Bello,
A.Battistoni,
G.Ricci,
J.Rossjohn,
H.O.Villar,
M.W.Parker.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The human pi-class glutathione S-transferase (hGST P1-1) is a target for
structure-based inhibitor design with the aim of developing drugs that could be
used as adjuvants in chemotherapeutic treatment. Here we present seven crystal
structures of the enzyme in complex with substrate (glutathione) and two
inhibitors (S-hexyl glutathione and gamma-glutamyl-
(S-benzyl)cysteinyl-D-phenylglycine). The binding of the modified glutathione
inhibitor, gamma-glutamyl-(S-benzyl)cysteinyl-D-phenylglycine, has been
characterized with the phenyl group stacking against the benzyl moiety of the
inhibitor and making interactions with the active-site residues Phe8 and Trp38.
The structure provides an explanation as to why this compound inhibits the
pi-class GST much better than the other GST classes. The structure of the enzyme
in complex with glutathione has been determined to high resolution (1.9 to 2.2
A) in three different crystal forms and at two different temperatures (100 and
288 K). In one crystal form, the direct hydrogen-bonding interaction between the
hydroxyl group of Tyr7, a residue involved in catalysis, and the thiol group of
the substrate, glutathione, is broken and replaced by a water molecule that
mediates the interaction. The hydrogen-bonding partner of the hydroxyl group of
Tyr108, another residue implicated in the catalysis, is space-group dependent. A
high-resolution (2.0 A) structure of the enzyme in complex with S-hexyl
glutathione in a new crystal form is presented. The enzyme-inhibitor complexes
show that the binding of ligand into the electrophilic binding site does not
lead to any conformational changes of the protein.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Schematic drawing of residues that interact with
the substrates and inhibitors. (a) GSH (C2 form, 100 K) and (b)
TER-117 (C2 form, 288 K). The key to the Figures is shown in
part (a). These Figures were produced using the program LIGPLOT
[Wallace et al 1995].
|
 |
Figure 7.
Figure 7. Superposition of the active sites of the
alpha-class hGST A1-1 [Sinning et al 1993], human mu-class GST
M2-2 [Raghunathan et al 1994] and human hGST P1-1 crystal
structures showing the fit of the inhibitor TER-117. The
structures are colored yellow, green and mauve, respectively.
The characteristic Mu-loop of the rat enzyme and the C-terminal
helix α9 of the alpha-class enzyme are shown. Residues likely
to collide with the phenyl ring of the inhibitor are
highlighted. This Figure was generated with the program package
INSIGHT II (Molecular Simulations Inc., San Diego, CA, USA.).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1997,
274,
84-0)
copyright 1997.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.F.Thévenin,
C.L.Zony,
B.J.Bahnson,
and
R.F.Colman
(2011).
GSTpi modulates JNK activity through a direct interaction with JNK substrate, ATF2.
|
| |
Protein Sci,
20,
834-848.
|
 |
|
|
|
|
 |
A.Oakley
(2011).
Glutathione transferases: a structural perspective.
|
| |
Drug Metab Rev,
43,
138-151.
|
 |
|
|
|
|
 |
G.McManus,
M.Costa,
A.Canals,
M.Coll,
and
T.J.Mantle
(2011).
Site-directed mutagenesis of mouse glutathione transferase P1-1 unlocks masked cooperativity, introduces a novel mechanism for 'ping pong' kinetic behaviour, and provides further structural evidence for participation of a water molecule in proton abstraction from glutathione.
|
| |
FEBS J,
278,
273-281.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Laborde
(2010).
Glutathione transferases as mediators of signaling pathways involved in cell proliferation and cell death.
|
| |
Cell Death Differ,
17,
1373-1380.
|
 |
|
|
|
|
 |
M.S.Goncalves,
J.P.Moura Neto,
C.L.Souza,
P.Melo,
and
M.G.Reis
(2010).
Evaluating glutathione S-transferase (GST) null genotypes (GSTT1 and GSTM1) as a potential biomarker of predisposition for developing leukopenia.
|
| |
Int J Lab Hematol,
32,
e49-e56.
|
 |
|
|
|
|
 |
V.T.Bhat,
A.M.Caniard,
T.Luksch,
R.Brenk,
D.J.Campopiano,
and
M.F.Greaney
(2010).
Nucleophilic catalysis of acylhydrazone equilibration for protein-directed dynamic covalent chemistry.
|
| |
Nat Chem,
2,
490-497.
|
 |
|
|
|
|
 |
J.Kaur,
and
A.K.Bachhawat
(2009).
Gln-222 in transmembrane domain 4 and Gln-526 in transmembrane domain 9 are critical for substrate recognition in the yeast high affinity glutathione transporter, Hgt1p.
|
| |
J Biol Chem,
284,
23872-23884.
|
 |
|
|
|
|
 |
D.F.Dourado,
P.A.Fernandes,
B.Mannervik,
and
M.J.Ramos
(2008).
Glutathione transferase: new model for glutathione activation.
|
| |
Chemistry,
14,
9591-9598.
|
 |
|
|
|
|
 |
R.E.Jenkins,
N.R.Kitteringham,
C.E.Goldring,
S.M.Dowdall,
J.Hamlett,
C.S.Lane,
J.S.Boerma,
N.P.Vermeulen,
and
B.K.Park
(2008).
Glutathione-S-transferase pi as a model protein for the characterisation of chemically reactive metabolites.
|
| |
Proteomics,
8,
301-315.
|
 |
|
|
|
|
 |
Y.C.Huang,
S.Misquitta,
S.Y.Blond,
E.Adams,
and
R.F.Colman
(2008).
Catalytically Active Monomer of Glutathione S-Transferase {pi} and Key Residues Involved in the Electrostatic Interaction between Subunits.
|
| |
J Biol Chem,
283,
32880-32888.
|
 |
|
|
|
|
 |
B.Blanchette,
X.Feng,
and
B.R.Singh
(2007).
Marine glutathione S-transferases.
|
| |
Mar Biotechnol (NY),
9,
513-542.
|
 |
|
|
|
|
 |
R.Téllez-Sanz,
E.Cesareo,
M.Nuccetelli,
A.M.Aguilera,
C.Barón,
L.J.Parker,
J.J.Adams,
C.J.Morton,
M.Lo Bello,
M.W.Parker,
and
L.García-Fuentes
(2006).
Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1.
|
| |
Protein Sci,
15,
1093-1105.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Cesareo,
L.J.Parker,
J.Z.Pedersen,
M.Nuccetelli,
A.P.Mazzetti,
A.Pastore,
G.Federici,
A.M.Caccuri,
G.Ricci,
J.J.Adams,
M.W.Parker,
and
M.Lo Bello
(2005).
Nitrosylation of human glutathione transferase P1-1 with dinitrosyl diglutathionyl iron complex in vitro and in vivo.
|
| |
J Biol Chem,
280,
42172-42180.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Cacciatore,
A.M.Caccuri,
A.Cocco,
F.De Maria,
A.Di Stefano,
G.Luisi,
F.Pinnen,
G.Ricci,
P.Sozio,
and
P.Turella
(2005).
Potent isozyme-selective inhibition of human glutathione S-transferase A1-1 by a novel glutathione S-conjugate.
|
| |
Amino Acids,
29,
255-261.
|
 |
|
|
|
|
 |
L.W.Yang,
and
I.Bahar
(2005).
Coupling between catalytic site and collective dynamics: a requirement for mechanochemical activity of enzymes.
|
| |
Structure,
13,
893-904.
|
 |
|
|
|
|
 |
M.Perbandt,
J.Höppner,
C.Betzel,
R.D.Walter,
and
E.Liebau
(2005).
Structure of the major cytosolic glutathione S-transferase from the parasitic nematode Onchocerca volvulus.
|
| |
J Biol Chem,
280,
12630-12636.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.A.Ralat,
and
R.F.Colman
(2004).
Glutathione S-transferase Pi has at least three distinguishable xenobiotic substrate sites close to its glutathione-binding site.
|
| |
J Biol Chem,
279,
50204-50213.
|
 |
|
|
|
|
 |
U.M.Hegazy,
B.Mannervik,
and
G.Stenberg
(2004).
Functional role of the lock and key motif at the subunit interface of glutathione transferase p1-1.
|
| |
J Biol Chem,
279,
9586-9596.
|
 |
|
|
|
|
 |
E.Ortiz-Salmerón,
M.Nuccetelli,
A.J.Oakley,
M.W.Parker,
M.Lo Bello,
and
L.García-Fuentes
(2003).
Thermodynamic description of the effect of the mutation Y49F on human glutathione transferase P1-1 in binding with glutathione and the inhibitor S-hexylglutathione.
|
| |
J Biol Chem,
278,
46938-46948.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.K.Kong,
G.Polekhina,
W.J.McKinstry,
M.W.Parker,
B.Dragani,
A.Aceto,
D.Paludi,
D.R.Principe,
B.Mannervik,
and
G.Stenberg
(2003).
Contribution of glycine 146 to a conserved folding module affecting stability and refolding of human glutathione transferase p1-1.
|
| |
J Biol Chem,
278,
1291-1302.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.M.Caccuri,
G.Antonini,
N.Allocati,
C.Di Ilio,
F.De Maria,
F.Innocenti,
M.W.Parker,
M.Masulli,
M.Lo Bello,
P.Turella,
G.Federici,
and
G.Ricci
(2002).
GSTB1-1 from Proteus mirabilis: a snapshot of an enzyme in the evolutionary pathway from a redox enzyme to a conjugating enzyme.
|
| |
J Biol Chem,
277,
18777-18784.
|
 |
|
|
|
|
 |
I.Le Trong,
R.E.Stenkamp,
C.Ibarra,
W.M.Atkins,
and
E.T.Adman
(2002).
1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
|
| |
Proteins,
48,
618-627.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Hirota,
and
Y.Hanyu
(2002).
Method for identification of mutant glutathione S-transferases conferring enhanced resistance to the anti-cancer drug chlorambucil.
|
| |
J Biosci Bioeng,
93,
618-621.
|
 |
|
|
|
|
 |
A.J.Oakley,
T.Harnnoi,
R.Udomsinprasert,
K.Jirajaroenrat,
A.J.Ketterman,
and
M.C.Wilce
(2001).
The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B.
|
| |
Protein Sci,
10,
2176-2185.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Polekhina,
P.G.Board,
A.C.Blackburn,
and
M.W.Parker
(2001).
Crystal structure of maleylacetoacetate isomerase/glutathione transferase zeta reveals the molecular basis for its remarkable catalytic promiscuity.
|
| |
Biochemistry,
40,
1567-1576.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Chang,
Y.G.Shin,
R.B.van Breemen,
S.Y.Blond,
and
J.L.Bolton
(2001).
Structural and functional consequences of inactivation of human glutathione S-transferase P1-1 mediated by the catechol metabolite of equine estrogens, 4-hydroxyequilenin.
|
| |
Biochemistry,
40,
4811-4820.
|
 |
|
|
|
|
 |
M.Lo Bello,
M.Nuccetelli,
A.M.Caccuri,
L.Stella,
M.W.Parker,
J.Rossjohn,
W.J.McKinstry,
A.F.Mozzi,
G.Federici,
F.Polizio,
J.Z.Pedersen,
and
G.Ricci
(2001).
Human glutathione transferase P1-1 and nitric oxide carriers; a new role for an old enzyme.
|
| |
J Biol Chem,
276,
42138-42145.
|
 |
|
|
|
|
 |
C.Micaloni,
A.P.Mazzetti,
M.Nuccetelli,
J.Rossjohn,
W.J.McKinstry,
G.Antonini,
A.M.Caccuri,
A.J.Oakley,
G.Federici,
G.Ricci,
M.W.Parker,
and
M.Lo Bello
(2000).
Valine 10 may act as a driver for product release from the active site of human glutathione transferase P1-1.
|
| |
Biochemistry,
39,
15961-15970.
|
 |
|
|
|
|
 |
L.Stella,
E.E.Di Iorio,
M.Nicotra,
and
G.Ricci
(1999).
Molecular dynamics simulations of human glutathione transferase P1-1: conformational fluctuations of the apo-structure.
|
| |
Proteins,
37,
10-19.
|
 |
|
|
|
|
 |
L.Stella,
M.Nicotra,
G.Ricci,
N.Rosato,
and
E.E.Di Iorio
(1999).
Molecular dynamics simulations of human glutathione transferase P1-1: analysis of the induced-fit mechanism by GSH binding.
|
| |
Proteins,
37,
1-9.
|
 |
|
|
|
|
 |
A.J.Oakley,
M.Lo Bello,
G.Ricci,
G.Federici,
and
M.W.Parker
(1998).
Evidence for an induced-fit mechanism operating in pi class glutathione transferases.
|
| |
Biochemistry,
37,
9912-9917.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Nicotra,
M.Paci,
M.Sette,
A.J.Oakley,
M.W.Parker,
M.Lo Bello,
A.M.Caccuri,
G.Federici,
and
G.Ricci
(1998).
Solution structure of glutathione bound to human glutathione transferase P1-1: comparison of NMR measurements with the crystal structure.
|
| |
Biochemistry,
37,
3020-3027.
|
 |
|
|
|
|
 |
R.N.Armstrong
(1998).
Mechanistic imperatives for the evolution of glutathione transferases.
|
| |
Curr Opin Chem Biol,
2,
618-623.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

