 |
PDBsum entry 8bgo
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
N,n-diacetylchitobiose deacetylase from pyrococcus chitonophagus with substrate n,n-diacetylchitobiose
|
|
Structure:
|
 |
Diacetylchitobiose deacetylase. Chain: a, h, k, b, e, l, g, c, i, j, f, d. Engineered: yes
|
|
Source:
|
 |
Thermococcus chitonophagus. Organism_taxid: 54262. Gene: a3l04_02905, chiton_0574. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
3.08Å
|
R-factor:
|
0.169
|
R-free:
|
0.215
|
|
|
Authors:
|
 |
W.Rypniewski,M.Bejger,K.Biniek-Antosiak
|
|
Key ref:
|
 |
K.Biniek-Antosiak
et al.
(2022).
Structural, Thermodynamic and enzymatic characterizat n , N -Diacetylchitobiose deacetylase from pyrococcus chitonophagus..
Int j mol sci,
23,
.
PubMed id:
|
 |
|
Date:
|
 |
|
28-Oct-22
|
Release date:
|
11-Jan-23
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A0A160VQZ8
(A0A160VQZ8_9EURY) -
Diacetylchitobiose deacetylase from Thermococcus chitonophagus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
267 a.a.
267 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.1.136
- N,N'-diacetylchitobiose non-reducing end deacetylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N,N'-diacetylchitobiose + H2O = beta-D-glucosaminyl-(1->4)-N-acetyl-D- glucosamine + acetate
|
 |
 |
 |
 |
 |
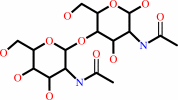 N,N'-diacetylchitobiose
N,N'-diacetylchitobiose
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 51.72% similarity
|
=
|
beta-D-glucosaminyl-(1->4)-N-acetyl-D- glucosamine
|
+
|
 acetate
acetate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

