 |
PDBsum entry 7uxt
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of ligand-free sethsa
|
|
Structure:
|
 |
Usg protein. Chain: b, a. Synonym: sethsa. Engineered: yes
|
|
Source:
|
 |
Streptococcus equi. Organism_taxid: 1336. Gene: gbl60_08830. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
3.40Å
|
R-factor:
|
0.238
|
R-free:
|
0.291
|
|
|
Authors:
|
 |
Y.Shi,V.Masic,T.Mosaiab,J.D.Nanson,B.Kobe,T.Ve
|
|
Key ref:
|
 |
m.k.manik
et al.
(2022).
Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science,
0,
eadc8969.
PubMed id:
|
 |
|
Date:
|
 |
|
06-May-22
|
Release date:
|
07-Sep-22
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
C0MAL8
(THSA_STRE4) -
NAD(+) hydrolase ThsA from Streptococcus equi subsp. equi (strain 4047)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
473 a.a.
467 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.2.5
- NAD(+) glycohydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
NAD+ + H2O = ADP-D-ribose + nicotinamide + H+
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
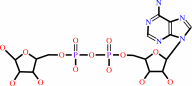 ADP-D-ribose
ADP-D-ribose
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Science
0:eadc8969
(2022)
|
|
PubMed id:
|
|
|
|

|
| |
|
Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
|
|
m.k.manik,
y.shi,
s.li,
m.a.zaydman,
n.damaraju,
s.eastmant.g.smith,
w.gu,
v.masic,
t.mosaiab,
j.s.weagley,
s.j.hance.vasquez,
l.hartley-tassell,
n.kargios,
n.maruta,
b.y.j.h.burdett,
m.j.landsberg,
m.a.schembri,
i.prokes,
l.song,
m.grant,
a.diantonio,
j.d.nanson,
m.guo,
j.milbrandt,
t.ve.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

