 |
PDBsum entry 7rtc
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of the arm domain from drosophila sarm1 in complex with namn
|
|
Structure:
|
 |
NAD(+) hydrolase sarm1. Chain: a, b, c. Synonym: nadase sarm1,sterile alpha and tir motif-containing protein 1,tir-1 homolog. Engineered: yes
|
|
Source:
|
 |
Drosophila melanogaster. Fruit fly. Organism_taxid: 7227. Gene: sarm, ect4, cg43119. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008
|
|
Resolution:
|
 |
|
3.31Å
|
R-factor:
|
0.239
|
R-free:
|
0.257
|
|
|
Authors:
|
 |
W.Gu,T.Ve,B.Kobe
|
|
Key ref:
|
 |
Y.Sasaki
et al.
(2021).
Nicotinic acid mononucleotide is an allosteric sarm1 inhibitor promoting axonal protection..
Exp neurol,
345,
13842.
PubMed id:
|
 |
|
Date:
|
 |
|
13-Aug-21
|
Release date:
|
01-Sep-21
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q6IDD9
(SARM1_DROME) -
NAD(+) hydrolase sarm1 from Drosophila melanogaster
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1360 a.a.
331 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.2.6
- ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
NAD+ + H2O = ADP-D-ribose + nicotinamide + H+
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 46.67% similarity
|
=
|
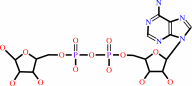 ADP-D-ribose
ADP-D-ribose
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

