 |
PDBsum entry 7rdf
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Flavoprotein
|
 |
|
Title:
|
 |
Crystal structure of pseudomonas aeruginosa d-arginine dehydrogenase y249f co-crystallized in the presence of d-arginine
|
|
Structure:
|
 |
Fad-dependent catabolic d-arginine dehydrogenase daua. Chain: a. Synonym: d-arginine dehydrogenase,dadh,d-arginine utilization protein a,dau. Engineered: yes
|
|
Source:
|
 |
Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1). Organism_taxid: 208964. Strain: atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1. Atcc: 15692. Gene: daua, pa3863. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.29Å
|
R-factor:
|
0.128
|
R-free:
|
0.157
|
|
|
Authors:
|
 |
R.A.G.Reis,A.Iyer,A.Agniswamy,I.T.Weber,G.Gadda
|
|
Key ref:
|
 |
A.Iyer
et al.
(2021).
Discovery of a new flavin n5-Adduct in a tyrosine to phenylalanine variant of d-Arginine dehydrogenase..
Arch.Biochem.Biophys.,
715,
09100.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
09-Jul-21
|
Release date:
|
22-Dec-21
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9HXE3
(DAUA_PSEAE) -
FAD-dependent catabolic D-arginine dehydrogenase DauA from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
375 a.a.
375 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.4.99.6
- D-arginine dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-arginine + A + H2O = 5-guanidino-2-oxopentanoate + AH2 + NH4+
|
 |
 |
 |
 |
 |
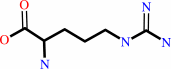 D-arginine
D-arginine
|
+
|
|
+
|
H2O
|
=
|
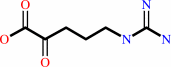 5-guanidino-2-oxopentanoate
5-guanidino-2-oxopentanoate
|
+
|
AH2
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
4R2)
matches with 85.48% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

