 |
PDBsum entry 7pim

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
7pim
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Partial structure of tyrosine hydroxylase lacking the first 35 residues in complex with dopamine.
|
|
Structure:
|
 |
Tyrosine 3-monooxygenase. Chain: b, a, d, f. Synonym: tyrosine 3-hydroxylase,th. Engineered: yes. Regulatory domain alpha-helix. Chain: g, c, e, h. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: th, tyh. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_taxid: 562
|
|
Authors:
|
 |
M.T.Bueno-Carrasco,J.Cuellar,C.Santiago,J.M.Valpuesta,A.Martinez, M.I.Flydal
|
|
Key ref:
|
 |
M.T.Bueno-Carrasco
et al.
(2022).
Structural mechanism for tyrosine hydroxylase inhibit dopamine and reactivation by ser40 phosphorylation..
Nat commun,
13,
74.
PubMed id:
|
 |
|
Date:
|
 |
|
20-Aug-21
|
Release date:
|
22-Dec-21
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains B, G, A, C, D, E, F, H:
E.C.1.14.16.2
- tyrosine 3-monooxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Dopa Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(6R)-L-erythro-5,6,7,8-tetrahydrobiopterin + L-tyrosine + O2 = (4aS,6R)- 4a-hydroxy-L-erythro-5,6,7,8-tetrahydrobiopterin + L-dopa
|
 |
 |
 |
 |
 |
(6R)-L-erythro-5,6,7,8-tetrahydrobiopterin
|
+
|
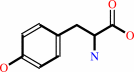 L-tyrosine
L-tyrosine
|
+
|
 O2
O2
|
=
|
(4aS,6R)- 4a-hydroxy-L-erythro-5,6,7,8-tetrahydrobiopterin
|
+
|
L-dopa
Bound ligand (Het Group name = )
matches with 78.57% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

