 |
PDBsum entry 7n2x

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
7n2x
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
The crystal structure of an fmn-dependent nadh:quinone oxidoreductase, azor from escherichia coli
|
|
Structure:
|
 |
Fmn-dependent nadh:quinone oxidoreductase. Chain: a. Synonym: azo-dye reductase,fmn-dependent nadh-azo compound oxidoreductase,fmn-dependent nadh-azoreductase. Engineered: yes
|
|
Source:
|
 |
Escherichia coli o157:h7. Organism_taxid: 83334. Gene: azor, z2315, ecs2014. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.167
|
R-free:
|
0.190
|
|
|
Authors:
|
 |
A.J.Arcinas,E.Fedorov,L.Kelly,S.C.Almo,A.Ghosh
|
|
Key ref:
|
 |
R.Hitchings
et al.
Uncovering a novel mechanism of enzyme activation in multimeric azoreductases.
To be published,
.
|
 |
|
Date:
|
 |
|
30-May-21
|
Release date:
|
31-Aug-22
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P41407
(AZOR_ECOLI) -
FMN-dependent NADH:quinone oxidoreductase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
201 a.a.
201 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.7.1.17
- FMN-dependent NADH-azoreductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N,N-dimethyl-1,4-phenylenediamine + anthranilate + 2 NAD+ = 2-(4-dimethylaminophenyl)diazenylbenzoate + 2 NADH + 2 H+
|
 |
 |
 |
 |
 |
N,N-dimethyl-1,4-phenylenediamine
Bound ligand (Het Group name = )
matches with 45.45% similarity
|
+
|
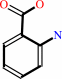 anthranilate
anthranilate
|
+
|
 2
×
NAD(+)
2
×
NAD(+)
|
=
|
2-(4-dimethylaminophenyl)diazenylbenzoate
|
+
|
 2
×
NADH
2
×
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FMN
|
 |
 |
 |
 |
 |
FMN
Bound ligand (Het Group name =
FMN)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

