 |
PDBsum entry 7mcp
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.5.1.48
- cystathionine gamma-synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
O-succinyl-L-homoserine + L-cysteine = L,L-cystathionine + succinate + H+
|
 |
 |
 |
 |
 |
O-succinyl-L-homoserine
Bound ligand (Het Group name = )
matches with 44.44% similarity
|
+
|
 L-cysteine
L-cysteine
|
=
|
L,L-cystathionine
|
+
|
 succinate
succinate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.4.4.1.1
- cystathionine gamma-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L,L-cystathionine + H2O = 2-oxobutanoate + L-cysteine + NH4+
|
 |
 |
 |
 |
 |
L,L-cystathionine
|
+
|
H2O
|
=
|
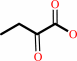 2-oxobutanoate
2-oxobutanoate
|
+
|
 L-cysteine
L-cysteine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.4.4.1.8
- Transferred entry: 4.4.1.13.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-cystathionine + H2O = L-homocysteine + NH3 + pyruvate
|
 |
 |
 |
 |
 |
 L-cystathionine
L-cystathionine
|
+
|
H(2)O
|
=
|
 L-homocysteine
L-homocysteine
|
+
|
NH(3)
|
+
|
 pyruvate
pyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Science
372:1169-1175
(2021)
|
|
PubMed id:
|
|
|
|

|
| |
|
Inhibitors of bacterial H2S biogenesis targeting antibiotic resistance and tolerance.
|
|
K.Shatalin,
A.Nuthanakanti,
A.Kaushik,
D.Shishov,
A.Peselis,
I.Shamovsky,
B.Pani,
M.Lechpammer,
N.Vasilyev,
E.Shatalina,
D.Rebatchouk,
A.Mironov,
P.Fedichev,
A.Serganov,
E.Nudler.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Emergent resistance to all clinical antibiotics calls for the next generation of
therapeutics. Here we report an effective antimicrobial strategy targeting the
bacterial hydrogen sulfide (H2S)-mediated defense system. We
identified cystathionine γ-lyase (CSE) as the primary generator of
H2S in two major human pathogens, Staphylococcus aureus and
Pseudomonas aeruginosa, and discovered small molecules that inhibit
bacterial CSE. These inhibitors potentiate bactericidal antibiotics against both
pathogens in vitro and in mouse models of infection. CSE inhibitors also
suppress bacterial tolerance, disrupting biofilm formation and substantially
reducing the number of persister bacteria that survive antibiotic treatment. Our
results establish bacterial H2S as a multifunctional defense factor
and CSE as a drug target for versatile antibiotic enhancers.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

