 |
PDBsum entry 7ekd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Biosynthetic protein
|
PDB id
|
|

|

|
7ekd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Biosynthetic protein
|
 |
|
Title:
|
 |
Crystal structure of gibberellin 3-oxidase 2 (ga3ox2) in rice
|
|
Structure:
|
 |
Gibberellin 3-beta-dioxygenase 2. Chain: a. Synonym: ga 3-oxidase 2,osga3ox2,gibberellin 3 beta-hydroxylase 2, protein dwarf18. Engineered: yes
|
|
Source:
|
 |
Oryza sativa subsp. Japonica. Rice. Organism_taxid: 39947. Gene: ga3ox2, d18, os01g0177400, loc_os01g08220, osj_00594, p0013f10.29. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.166
|
R-free:
|
0.212
|
|
|
Authors:
|
 |
S.Takehara,K.Kawai,B.Mikami,M.Ueguchi-Tanaka
|
|
Key ref:
|
 |
K.Kawai
et al.
(2022).
Evolutionary alterations in gene expression and enzym activities of gibberellin 3-Oxidase 1 in oryza..
Commun biol,
5,
67.
PubMed id:
|
 |
|
Date:
|
 |
|
05-Apr-21
|
Release date:
|
16-Feb-22
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9FU53
(G3OX2_ORYSJ) -
Gibberellin 3-beta-dioxygenase 2 from Oryza sativa subsp. japonica
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
373 a.a.
343 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.11.15
- gibberellin 3beta-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Diterpenoid biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
gibberellin A20 + 2-oxoglutarate + O2 = gibberellin A1 + succinate + CO2
|
 |
 |
 |
 |
 |
gibberellin A20
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
2-oxoglutarate
Bound ligand (Het Group name = )
matches with 95.83% similarity
|
+
|
 O2
O2
|
=
|
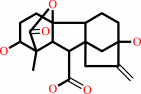 gibberellin A1
gibberellin A1
|
+
|
 succinate
succinate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(2+); L-ascorbate
|
 |
 |
 |
 |
 |
Fe(2+)
|
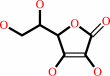 L-ascorbate
L-ascorbate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

