 |
PDBsum entry 7ch3

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 469 a.a.
469 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 5 more)
495 a.a.
(+ 5 more)
495 a.a.
|
 |

|

|
|
|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of arabinose isomerase from hyper thermophilic bacterium thermotoga maritima (tmai) triple mutant (k264a, e265a, k266a)
|
|
Structure:
|
 |
L-arabinose isomerase. Chain: a, b, c, d, e, f, g, h, i, j, k, l. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099). Organism_taxid: 243274. Gene: araa, tm_0276. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008
|
|
Resolution:
|
 |
|
3.61Å
|
R-factor:
|
0.234
|
R-free:
|
0.280
|
|
|
Authors:
|
 |
T.P.Cao,I.Dhanasingh,J.Y.Sung,S.M.Shin,D.W.Lee,S.H.Lee
|
|
Key ref:
|
 |
T.P.Cao
et al.
Crystal structure of arabinose isomerase from hyper thermophilic bacterium thermotoga maritima (tmai) tri mutant (k264a, E265a, K266a).
To be published,
.
|
 |
|
Date:
|
 |
|
04-Jul-20
|
Release date:
|
20-Oct-21
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L:
E.C.5.3.1.4
- L-arabinose isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-L-arabinopyranose = L-ribulose
|
 |
 |
 |
 |
 |
beta-L-arabinopyranose
|
=
|
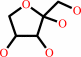 L-ribulose
L-ribulose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

