 |
PDBsum entry 7ba4

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Cytosolic protein
|
PDB id
|
|

|

|
7ba4
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Cytosolic protein
|
 |
|
Title:
|
 |
Structure of cystathionine gamma-lyase from pseudomonas aeruginosa
|
|
Structure:
|
 |
Cystathionine gamma-lyase. Chain: a, b, c, d. Synonym: cystathionine gamma-synthase. Engineered: yes
|
|
Source:
|
 |
Pseudomonas aeruginosa. Organism_taxid: 287. Gene: cgu42_31115, ecc04_012645, fc629_02215, gnq48_07915, ipc582_10875. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.162
|
R-free:
|
0.205
|
|
|
Authors:
|
 |
C.Fernandez-Rodriguez,I.Oyenarte,I.Gonzalez-Recio,T.Majtan,A.Astegno, L.A.Martinez-Cruz
|
|
Key ref:
|
 |
C.Fernandez-Rodriguez
et al.
Structure of cystathionine gamma-Lyase from pseudomon aeruginosa.
To be published,
.
|
 |
|
Date:
|
 |
|
15-Dec-20
|
Release date:
|
29-Sep-21
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D:
E.C.2.5.1.48
- cystathionine gamma-synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
O-succinyl-L-homoserine + L-cysteine = L,L-cystathionine + succinate + H+
|
 |
 |
 |
 |
 |
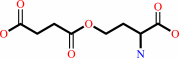 O-succinyl-L-homoserine
O-succinyl-L-homoserine
|
+
|
 L-cysteine
L-cysteine
|
=
|
L,L-cystathionine
|
+
|
 succinate
succinate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

