 |
PDBsum entry 6vue

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lyase/inhibitor
|
PDB id
|
|

|

|
6vue
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase/inhibitor
|
 |
|
Title:
|
 |
Wild-type choline tma lyase in complex with 1-methyl-1,2,3,6- tetrahydropyridin-3-ol
|
|
Structure:
|
 |
Choline trimethylamine-lyase. Chain: a, b. Synonym: choline tma-lyase,choline utilization protein c,glycyl radical enzyme cutc,gre cutc. Engineered: yes
|
|
Source:
|
 |
Desulfovibrio alaskensis (strain g20). Organism_taxid: 207559. Strain: g20. Gene: cutc, dde_3282. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008. Expression_system_variant: rosetta 2
|
|
Resolution:
|
 |
|
2.28Å
|
R-factor:
|
0.165
|
R-free:
|
0.197
|
|
|
Authors:
|
 |
M.A.Ortega,C.L.Drennan
|
|
Key ref:
|
 |
M.Bollenbach
et al.
(2020).
Discovery of a Cyclic Choline Analog That Inhibits Anaerobic Choline Metabolism by Human Gut Bacteria.
ACS Med Chem Lett,
11,
1980-1985.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
15-Feb-20
|
Release date:
|
04-Nov-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q30W70
(CUTC_DESAG) -
Choline trimethylamine-lyase from Oleidesulfovibrio alaskensis (strain ATCC BAA-1058 / DSM 17464 / G20)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
846 a.a.
803 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 7 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.99.4
- choline trimethylamine-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
choline = trimethylamine + acetaldehyde
|
 |
 |
 |
 |
 |
choline
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
=
|
 trimethylamine
trimethylamine
|
+
|
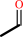 acetaldehyde
acetaldehyde
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
ACS Med Chem Lett
11:1980-1985
(2020)
|
|
PubMed id:
|
|
|
|

|
| |
|
Discovery of a Cyclic Choline Analog That Inhibits Anaerobic Choline Metabolism by Human Gut Bacteria.
|
|
M.Bollenbach,
M.Ortega,
M.Orman,
C.L.Drennan,
E.P.Balskus.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The anaerobic conversion of choline to trimethylamine (TMA) by the human gut
microbiota has been linked to multiple human diseases. The potential impact of
this microbial metabolic activity on host health has inspired multiple efforts
to identify small molecule inhibitors. Here, we use information about the
structure and mechanism of the bacterial enzyme choline TMA-lyase (CutC) to
develop a cyclic choline analog that inhibits the conversion of choline to TMA
in bacterial whole cells and in a complex gut microbial community. In
vitro biochemical assays and a crystal structure suggest that this analog is
a competitive, mechanism-based inhibitor. This work demonstrates the utility of
structure-based design to access inhibitors of radical enzymes from the human
gut microbiota.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

