 |
PDBsum entry 6t3t
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Structure of the 4-hydroxy-tetrahydrodipicolinate synthase from the thermoacidophilic methanotroph methylacidiphilum fumariolicum solv
|
|
Structure:
|
 |
4-hydroxy-tetrahydrodipicolinate synthase. Chain: b, a, c, d. Synonym: htpa synthase. Ec: 4.3.3.7
|
|
Source:
|
 |
Methylacidiphilum fumariolicum solv. Organism_taxid: 1156937
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.184
|
R-free:
|
0.232
|
|
|
Authors:
|
 |
R.Schmitz,A.Dietl,M.Mueller,T.Berben,H.Op Den Camp,T.Barends
|
|
Key ref:
|
 |
R.A.Schmitz
et al.
(2020).
Structure of the 4-hydroxy-tetrahydrodipicolinate synthase from the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV and the phylogeny of the aminotransferase pathway.
Acta Crystallogr F Struct Biol Commun,
76,
199-208.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
11-Oct-19
|
Release date:
|
06-May-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
I0JZ23
(I0JZ23_METFB) -
4-hydroxy-tetrahydrodipicolinate synthase from Methylacidiphilum fumariolicum (strain SolV)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
301 a.a.
300 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.3.7
- 4-hydroxy-tetrahydrodipicolinate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate 4-semialdehyde + pyruvate = (2S,4S)-4-hydroxy-2,3,4,5- tetrahydrodipicolinate + H2O + H+
|
 |
 |
 |
 |
 |
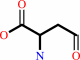 L-aspartate 4-semialdehyde
L-aspartate 4-semialdehyde
|
+
|
 pyruvate
pyruvate
|
=
|
(2S,4S)-4-hydroxy-2,3,4,5- tetrahydrodipicolinate
|
+
|
H2O
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr F Struct Biol Commun
76:199-208
(2020)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of the 4-hydroxy-tetrahydrodipicolinate synthase from the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV and the phylogeny of the aminotransferase pathway.
|
|
R.A.Schmitz,
A.Dietl,
M.Müller,
T.Berben,
H.J.M.Op den Camp,
T.R.M.Barends.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The enzyme 4-hydroxy-tetrahydrodipicolinate synthase (DapA) is involved in the
production of lysine and precursor molecules for peptidoglycan synthesis. In a
multistep reaction, DapA converts pyruvate and L-aspartate-4-semialdehyde to
4-hydroxy-2,3,4,5-tetrahydrodipicolinic acid. In many organisms, lysine binds
allosterically to DapA, causing negative feedback, thus making the enzyme an
important regulatory component of the pathway. Here, the 2.1 Å resolution
crystal structure of DapA from the thermoacidophilic methanotroph
Methylacidiphilum fumariolicum SolV is reported. The enzyme crystallized as a
contaminant of a protein preparation from native biomass. Genome analysis
reveals that M. fumariolicum SolV utilizes the recently discovered
aminotransferase pathway for lysine biosynthesis. Phylogenetic analyses of the
genes involved in this pathway shed new light on the distribution of this
pathway across the three domains of life.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

