 |
PDBsum entry 6t37
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Pseudomonas aeruginosa rmla in complex with allosteric inhibitor
|
|
Structure:
|
 |
Glucose-1-phosphate thymidylyltransferase. Chain: a, b, c, d. Engineered: yes. Other_details: residues missing from the model are due to flexibility and weak electron density
|
|
Source:
|
 |
Pseudomonas aeruginosa. Organism_taxid: 287. Gene: rmla, rfba, caz10_00575, e4v10_15605, ipc1492_24235, ipc605_02475, pamh19_2921. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.08Å
|
R-factor:
|
0.219
|
R-free:
|
0.242
|
|
|
Authors:
|
 |
M.S.Alphey,G.Xiao,J.N.Westwood
|
|
Key ref:
|
 |
G.Xiao
et al.
Potent allosteric inhibitors of pseudomonas aeruginos glucose-1-Phosphate thymidylyltransferase (rmla).
To be published,
.
PubMed id:
|
 |
|
Date:
|
 |
|
10-Oct-19
|
Release date:
|
19-Aug-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9HU22
(Q9HU22_PSEAE) -
Glucose-1-phosphate thymidylyltransferase from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
293 a.a.
289 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.24
- glucose-1-phosphate thymidylyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
6-Deoxyhexose Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dTTP + alpha-D-glucose 1-phosphate + H+ = dTDP-alpha-D-glucose + diphosphate
|
 |
 |
 |
 |
 |
 dTTP
dTTP
|
+
|
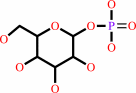 alpha-D-glucose 1-phosphate
alpha-D-glucose 1-phosphate
|
+
|
H(+)
|
=
|
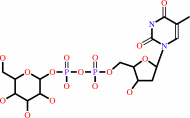 dTDP-alpha-D-glucose
dTDP-alpha-D-glucose
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

