 |
PDBsum entry 6t0b

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
6t0b
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 431 a.a.
431 a.a.
|
 |

|

|

|

|

|

|

|

 352 a.a.
352 a.a.
|
 |

|

|

|

|

|

|

|

 385 a.a.
385 a.a.
|
 |

|

|

|

|

|

|

|

 247 a.a.
247 a.a.
|
 |

|

|

|

|

|

|

|

 185 a.a.
185 a.a.
|
 |

|

|

|

|

|

|

|

 75 a.a.
75 a.a.
|
 |

|

|

|

|

|

|

|

 126 a.a.
126 a.a.
|
 |

|

|

|

|

|

|

|

 93 a.a.
93 a.a.
|
 |

|

|

|

|

|

|

|

 57 a.a.
57 a.a.
|
 |

|

|

|

|

|

|

|

 76 a.a.
76 a.a.
|
 |

|

|

|

|

|

|

|

 534 a.a.
534 a.a.
|
 |

|

|

|

|

|

|

|

 236 a.a.
236 a.a.
|
 |

|

|

|

|

|

|

|

 269 a.a.
269 a.a.
|
 |

|

|

|

|

|

|

|

 120 a.a.
120 a.a.
|
 |

|

|

|

|

|

|

|

 133 a.a.
133 a.a.
|
 |

|

|

|

|

|

|

|

 102 a.a.
102 a.a.
|
 |

|

|

|

|

|

|

|

 59 a.a.
59 a.a.
|
 |

|

|

|

|

|

|

|

 51 a.a.
51 a.a.
|
 |

|

|

|

|

|

|

|

 55 a.a.
55 a.a.
|
 |

|

|

|

|

|

|

|

 75 a.a.
75 a.a.
|
 |

|

|

|

|

|

|

|

 113 a.a.
113 a.a.
|
 |

|

|

|

|

|

|

|

 45 a.a.
45 a.a.
|
 |

|

|

|

|

|

|

|

 99 a.a.
99 a.a.
|
 |

|

|
|
|
|

|

|

|

|

|
 _ZN
×2 _ZN
×2
|
 |

|

|

|

|

|

|

|
 _CU
×2 _CU
×2
|
 |

|

|

|

|

|

|

|
 _CA
×2 _CA
×2
|
 |

|

|

|

|

|

|

|
 _MG
×2 _MG
×2
|
 |

|

|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
The iii2-iv(5b)2 respiratory supercomplex from s. Cerevisiae
|
|
Structure:
|
 |
Cytochrome b-c1 complex subunit 1, mitochondrial. Chain: a, l. Synonym: complex iii subunit 1,core protein i,ubiquinol-cytochromE-C reductase complex core protein 1. Cytochrome b-c1 complex subunit 2, mitochondrial. Chain: b, m. Synonym: complex iii subunit 2,core protein ii,ubiquinol-cytochromE-C reductase complex core protein 2. Cytochrome b.
|
|
Source:
|
 |
Saccharomyces cerevisiae s288c. Baker's yeast. Organism_taxid: 559292. Gene: cox13, ygl191w, g1341. Expressed in: saccharomyces cerevisiae s288c. Expression_system_taxid: 559292. Expression_system_variant: w303-1b. Organism_taxid: 559292
|
|
Authors:
|
 |
A.Marechal,N.Pinotsis,A.Hartley
|
|
Key ref:
|
 |
A.M.Hartley
et al.
(2020).
Rcf2 revealed in cryo-EM structures of hypoxic isoforms of mature mitochondrial III-IV supercomplexes.
Proc Natl Acad Sci U S A,
117,
9329-9337.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
02-Oct-19
|
Release date:
|
22-Apr-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P07256
(QCR1_YEAST) -
Cytochrome b-c1 complex subunit 1, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
457 a.a.
431 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P07257
(QCR2_YEAST) -
Cytochrome b-c1 complex subunit 2, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
368 a.a.
352 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00163
(CYB_YEAST) -
Cytochrome b from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
385 a.a.
385 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P07143
(CY1_YEAST) -
Cytochrome c1, heme protein, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
309 a.a.
247 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P08067
(UCRI_YEAST) -
Cytochrome b-c1 complex subunit Rieske, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
215 a.a.
185 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00127
(QCR6_YEAST) -
Cytochrome b-c1 complex subunit 6, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
147 a.a.
75 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00128
(QCR7_YEAST) -
Cytochrome b-c1 complex subunit 7, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
127 a.a.
126 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P08525
(QCR8_YEAST) -
Cytochrome b-c1 complex subunit 8, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
94 a.a.
93 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P22289
(QCR9_YEAST) -
Cytochrome b-c1 complex subunit 9, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
66 a.a.
57 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P37299
(QCR10_YEAST) -
Cytochrome b-c1 complex subunit 10, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
77 a.a.
76 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00401
(COX1_YEAST) -
Cytochrome c oxidase subunit 1 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
534 a.a.
534 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00410
(COX2_YEAST) -
Cytochrome c oxidase subunit 2 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
251 a.a.
236 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00420
(COX3_YEAST) -
Cytochrome c oxidase subunit 3 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
269 a.a.
269 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P04037
(COX4_YEAST) -
Cytochrome c oxidase subunit 4, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
155 a.a.
120 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00425
(COX5B_YEAST) -
Cytochrome c oxidase subunit 5B, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
151 a.a.
133 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00427
(COX6_YEAST) -
Cytochrome c oxidase subunit 6, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
148 a.a.
102 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P10174
(COX7_YEAST) -
Cytochrome c oxidase subunit 7, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
60 a.a.
59 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P04039
(COX8_YEAST) -
Cytochrome c oxidase subunit 8, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
78 a.a.
51 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P07255
(COX9_YEAST) -
Cytochrome c oxidase subunit 9, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
59 a.a.
55 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
Q01519
(COX12_YEAST) -
Cytochrome c oxidase subunit 12, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
83 a.a.
75 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P32799
(COX13_YEAST) -
Cytochrome c oxidase subunit 13, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
129 a.a.
113 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains a, b, c, n, o, p:
E.C.7.1.1.9
- cytochrome-c oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
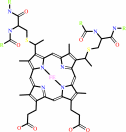
4 Fe(II)-[cytochrome c] + O2 + 8 H+(in) = 4 Fe(III)-[cytochrome c] + 2 H2O + 4 H+(out)
|
 |
 |
 |
 |
 |
4
×
Fe(II)-[cytochrome c]
|
+
|
 O2
O2
|
+
|
8
×
H(+)(in)
|
=
|
4
×
Fe(III)-[cytochrome c]
|
+
|
2
×
H2O
|
+
|
4
×
H(+)(out)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains C, D, E, N, O, P:
E.C.7.1.1.8
- quinol--cytochrome-c reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a quinol + 2 Fe(III)-[cytochrome c](out) = a quinone + 2 Fe(II)- [cytochrome c](out) + 2 H(+)(out)
|
 |
 |
 |
 |
 |
4
×
quinol
Bound ligand (Het Group name = )
matches with 63.64% similarity
|
+
|
2
×
Fe(III)-[cytochrome c](out)
|
=
|
 8
×
quinone
8
×
quinone
|
+
|
2
×
Fe(II)- [cytochrome c](out)
|
+
|
2
×
H(+)(out)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chains d, e, f, g, h, i, j, k, q, r, s, t, u, v, w, x:
E.C.1.9.3.1
- Transferred entry: 7.1.1.9.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4 ferrocytochrome c + O2 + 4 H+ = 4 ferricytochrome c + 2 H2O
|
 |
 |
 |
 |
 |
 4
×
ferrocytochrome c
4
×
ferrocytochrome c
|
+
|
 2
×
O(2)
2
×
O(2)
|
+
|
4
×
H(+)
|
=
|
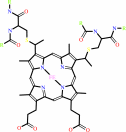 4
×
ferricytochrome c
4
×
ferricytochrome c
|
+
|
2
×
H(2)O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
117:9329-9337
(2020)
|
|
PubMed id:
|
|
|
|

|
| |
|
Rcf2 revealed in cryo-EM structures of hypoxic isoforms of mature mitochondrial III-IV supercomplexes.
|
|
A.M.Hartley,
B.Meunier,
N.Pinotsis,
A.Maréchal.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The organization of the mitochondrial electron transport chain proteins into
supercomplexes (SCs) is now undisputed; however, their assembly process, or the
role of differential expression isoforms, remain to be determined. In
Saccharomyces cerevisiae, cytochrome c oxidase (CIV) forms SCs of
varying stoichiometry with cytochrome bc1 (CIII). Recent
studies have revealed, in normoxic growth conditions, an interface made
exclusively by Cox5A, the only yeast respiratory protein that exists as one of
two isoforms depending on oxygen levels. Here we present the cryo-EM structures
of the III2-IV1 and III2-IV2 SCs
containing the hypoxic isoform Cox5B solved at 3.4 and 2.8 Å, respectively. We
show that the change of isoform does not affect SC formation or activity, and
that SC stoichiometry is dictated by the level of CIII/CIV biosynthesis.
Comparison of the CIV5B- and CIV5A-containing SC
structures highlighted few differences, found mainly in the region of Cox5.
Additional density was revealed in all SCs, independent of the CIV isoform, in a
pocket formed by Cox1, Cox3, Cox12, and Cox13, away from the CIII-CIV interface.
In the CIV5B-containing hypoxic SCs, this could be confidently
assigned to the hypoxia-induced gene 1 (Hig1) type 2 protein Rcf2. With
conserved residues in mammalian Hig1 proteins and Cox3/Cox12/Cox13 orthologs, we
propose that Hig1 type 2 proteins are stoichiometric subunits of CIV, at least
when within a III-IV SC.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |

| | |

