 |
PDBsum entry 6o5h
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.3.2.6
- leukotriene-A4 hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
leukotriene A4 + H2O = leukotriene B4
|
 |
 |
 |
 |
 |
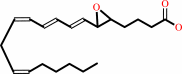 leukotriene A4
leukotriene A4
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 41.94% similarity
|
=
|
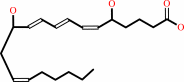 leukotriene B4
leukotriene B4
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.4.11.4
- tripeptide aminopeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Release of a N-terminal residue from a tripeptide.
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Med Chem
62:10605-10616
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
Effect of Modifier Structure on the Activation of Leukotriene A4 Hydrolase Aminopeptidase Activity.
|
|
K.H.Lee,
G.Petruncio,
A.Shim,
M.Burdick,
Z.Zhang,
Y.M.Shim,
S.M.Noble,
M.Paige.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Activation of the leukotriene A4 hydrolase (LTA4H)
aminopeptidase (AP) activity with 4-methoxydiphenylmethane (4MDM) promoted
resolution of neutrophil infiltration in a murine cigarette smoke-induced model
for emphysematous chronic obstructive pulmonary disease. Recently,
4-(4-benzylphenyl)thiazol-2-amine (ARM1) was published as a ligand for
LTA4H with potential anti-inflammatory properties. To investigate the
effect of modifier structure on enzyme kinetics of LTA4H, a series of
analogues bearing structural features of ARM1 and 4MDM were synthesized using
trifluoroborate Suzuki coupling reactions. Following, the 2.8 Å X-ray crystal
structure of LTA4H complexed with 4-OMe-ARM1, a 4MDM-ARM1 hybrid
molecule, was determined. Kinetic analysis showed that ARM1 and related
analogues lowered affinity for the enzyme-substrate complex, resulting in a
change of mechanism from hyperbolic mixed predominately catalytic activation
(HMx(Sp < Ca)A) as observed for 4MDM to a predominately specific activation
(HMx(Sp > Ca)A) mechanism. 4-OMe-ARM1 was then shown to dose responsively
reduce LTB4 production in human neutrophils.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

