 |
PDBsum entry 6ecr

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
6ecr
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.5.1.5
- methylenetetrahydrofolate dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Folate Coenzymes
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(6R)-5,10-methylene-5,6,7,8-tetrahydrofolate + NADP+ = (6R)-5,10- methenyltetrahydrofolate + NADPH
|
 |
 |
 |
 |
 |
(6R)-5,10-methylene-5,6,7,8-tetrahydrofolate
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
(6R)-5,10- methenyltetrahydrofolate
|
+
|
 NADPH
NADPH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.5.4.9
- methenyltetrahydrofolate cyclohydrolase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(6R)-5,10-methenyltetrahydrofolate + H2O = (6R)-10-formyltetrahydrofolate + H+
|
 |
 |
 |
 |
 |
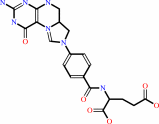 5,10-methenyltetrahydrofolate
5,10-methenyltetrahydrofolate
|
+
|
H2O
|
=
|
(6S)-10-formyltetrahydrofolate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.6.3.4.3
- formate--tetrahydrofolate ligase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(6S)-5,6,7,8-tetrahydrofolate + formate + ATP = (6R)-10- formyltetrahydrofolate + ADP + phosphate
|
 |
 |
 |
 |
 |
(6S)-5,6,7,8-tetrahydrofolate
|
+
|
 formate
formate
|
+
|
ATP
Bound ligand (Het Group name = )
matches with 75.00% similarity
|
=
|
(6R)-10- formyltetrahydrofolate
|
+
|
ADP
Bound ligand (Het Group name = )
matches with 56.25% similarity
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr F Struct Biol Commun
75:148-152
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
An assessment of three human methylenetetrahydrofolate dehydrogenase/cyclohydrolase-ligand complexes following further refinement.
|
|
R.Bueno,
A.Dawson,
W.N.Hunter.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The enzymes involved in folate metabolism are key drug targets for cell-growth
modulation, and accurate crystallographic structures provide templates to be
exploited for structure-based ligand design. In this context, three ternary
complex structures of human methylenetetrahydrofolate
dehydrogenase/cyclohydrolase have been published [Schmidt et al. (2000),
Biochemistry, 39, 6325-6335] and potentially represent starting points for the
development of new antifolate inhibitors. However, an inspection of the models
and the deposited data revealed deficiencies and raised questions about the
validity of the structures. A number of inconsistencies relating to the
publication were also identified. Additional refinement was carried out with the
deposited data, seeking to improve the models and to then validate the complex
structures or correct the record. In one case, the inclusion of the inhibitor in
the structure was supported and alterations to the model allowed details of
enzyme-ligand interactions to be described that had not previously been
discussed. For one weak inhibitor, the data suggested that the ligand may adopt
two poses in the binding site, both with few interactions with the enzyme. In
the third case, that of a potent inhibitor, inconsistencies were noted in the
assignment of the chemical structure and there was no evidence to support the
inclusion of the ligand in the active site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

