 |
PDBsum entry 6drh
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Toxin
|
 |
|
Title:
|
 |
Adp-ribosyltransferase toxin/immunity pair
|
|
Structure:
|
 |
Adp-ribosyl-(dinitrogen reductase) hydrolase. Chain: a, c, e, g. Engineered: yes. Paar repeat-containing protein. Chain: b, d, f, h. Engineered: yes
|
|
Source:
|
 |
Serratia proteamaculans (strain 568). Organism_taxid: 399741. Strain: 568. Gene: spro_3018. Expressed in: escherichia coli. Expression_system_taxid: 562. Gene: spro_3017. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.153
|
R-free:
|
0.206
|
|
|
Authors:
|
 |
D.E.Bosch,S.Ting,M.Allaire,J.D.Mougous
|
|
Key ref:
|
 |
S.Y.Ting
et al.
(2018).
Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins.
Cell,
175,
1380.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
11-Jun-18
|
Release date:
|
31-Oct-18
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, C, E, G:
E.C.3.2.2.19
- [protein ADP-ribosylarginine] hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
N(omega)-(ADP-D-ribosyl)-L-arginyl-[protein] + H2O = ADP-D-ribose + L-arginyl-[protein]
|
|
2.
|
N(omega)-(ADP-D-ribosyl)-L-arginine + H2O = ADP-D-ribose + L-arginine
|
|
 |
 |
 |
 |
 |
N(omega)-(ADP-D-ribosyl)-L-arginyl-[protein]
|
+
|
H2O
|
=
|
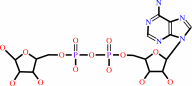 ADP-D-ribose
ADP-D-ribose
|
+
|
L-arginyl-[protein]
|
|
 |
 |
 |
 |
 |
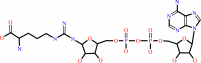 N(omega)-(ADP-D-ribosyl)-L-arginine
N(omega)-(ADP-D-ribosyl)-L-arginine
|
+
|
H2O
|
=
|
 ADP-D-ribose
ADP-D-ribose
|
+
|
 L-arginine
L-arginine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains B, D, F, H:
E.C.2.4.2.31
- NAD(+)--protein-arginine ADP-ribosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-arginyl-[protein] + NAD+ = N(omega)-(ADP-D-ribosyl)-L-arginyl- [protein] + nicotinamide + H+
|
 |
 |
 |
 |
 |
L-arginyl-[protein]
|
+
|
 NAD(+)
NAD(+)
|
=
|
N(omega)-(ADP-D-ribosyl)-L-arginyl- [protein]
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Cell
175:1380
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins.
|
|
S.Y.Ting,
D.E.Bosch,
S.M.Mangiameli,
M.C.Radey,
S.Huang,
Y.J.Park,
K.A.Kelly,
S.K.Filip,
Y.A.Goo,
J.K.Eng,
M.Allaire,
D.Veesler,
P.A.Wiggins,
S.B.Peterson,
J.D.Mougous.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
ADP-ribosylation of proteins can profoundly impact their function and serves as
an effective mechanism by which bacterial toxins impair eukaryotic cell
processes. Here, we report the discovery that bacteria also employ
ADP-ribosylating toxins against each other during interspecies competition. We
demonstrate that one such toxin from Serratia proteamaculans interrupts the
division of competing cells by modifying the essential bacterial tubulin-like
protein, FtsZ, adjacent to its protomer interface, blocking its capacity to
polymerize. The structure of the toxin in complex with its immunity determinant
revealed two distinct modes of inhibition: active site occlusion and enzymatic
removal of ADP-ribose modifications. We show that each is sufficient to support
toxin immunity; however, the latter additionally provides unprecedented broad
protection against non-cognate ADP-ribosylating effectors. Our findings reveal
how an interbacterial arms race has produced a unique solution for safeguarding
the integrity of bacterial cell division machinery against inactivating
post-translational modifications.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

