 |
PDBsum entry 6ddc
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of the single mutant (d52n) of nt5c2-537x in the basal state, northeast structural genomics consortium target
|
|
Structure:
|
 |
Cytosolic purine 5'-nucleotidase. Chain: a, b. Fragment: residues 1-536. Synonym: cytosolic 5'-nucleotidase ii. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: nt5c2, nt5b, nt5cp, pnt5. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_cell: rosetta 2(de3).
|
|
Resolution:
|
 |
|
2.91Å
|
R-factor:
|
0.187
|
R-free:
|
0.264
|
|
|
Authors:
|
 |
F.Forouhar,C.L.Dieck,G.Tzoneva,Z.Carpenter,A.Ambesi-Impiombato, M.Sanchez-Martin,R.Kirschner-Schwabe,S.Lew,J.Seetharaman, A.A.Ferrando,L.Tong,Northeast Structural Genomics Consortium (Nesg)
|
|
Key ref:
|
 |
C.L.Dieck
et al.
(2018).
Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell,
34,
136.
PubMed id:
|
 |
|
Date:
|
 |
|
09-May-18
|
Release date:
|
04-Jul-18
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P49902
(5NTC_HUMAN) -
Cytosolic purine 5'-nucleotidase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
561 a.a.
475 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.1.77
- nucleoside phosphotransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2'-deoxyribonucleoside + a ribonucleoside 5'-phosphate = a ribonucleoside + a 2'-deoxyribonucleoside 5'-phosphate
|
 |
 |
 |
 |
 |
2'-deoxyribonucleoside
|
+
|
ribonucleoside 5'-phosphate
|
=
|
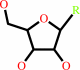 ribonucleoside
ribonucleoside
|
+
|
2'-deoxyribonucleoside 5'-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.3.5
- 5'-nucleotidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a ribonucleoside 5'-phosphate + H2O = a ribonucleoside + phosphate
|
 |
 |
 |
 |
 |
ribonucleoside 5'-phosphate
|
+
|
H2O
|
=
|
 ribonucleoside
ribonucleoside
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.1.3.99
- IMP-specific 5'-nucleotidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + H2O = inosine + phosphate
|
 |
 |
 |
 |
 |
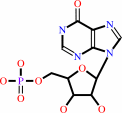 IMP
IMP
|
+
|
H2O
|
=
|
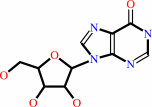 inosine
inosine
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent metal cation
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Cancer Cell
34:136
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
|
|
C.L.Dieck,
G.Tzoneva,
F.Forouhar,
Z.Carpenter,
A.Ambesi-Impiombato,
M.Sánchez-Martín,
R.Kirschner-Schwabe,
S.Lew,
J.Seetharaman,
L.Tong,
A.A.Ferrando.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Activating mutations in the cytosolic 5'-nucleotidase II gene NT5C2 drive
resistance to 6-mercaptopurine in acute lymphoblastic leukemia. Here we
demonstrate that constitutively active NT5C2 mutations K359Q and L375F
reconfigure the catalytic center for substrate access and catalysis in the
absence of allosteric activator. In contrast, most relapse-associated mutations,
which involve the arm segment and residues along the surface of the
inter-monomeric cavity, disrupt a built-in switch-off mechanism responsible for
turning off NT5C2. In addition, we show that the C-terminal acidic tail lost in
the Q523X mutation functions to restrain NT5C2 activation. These results uncover
dynamic mechanisms of enzyme regulation targeted by chemotherapy
resistance-driving NT5C2 mutations, with important implications for the
development of NT5C2 inhibitor therapies.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

