 |
PDBsum entry 6afb
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.2.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.5.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.5.1.124
- protein deglycase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
N(omega)-(1-hydroxy-2-oxopropyl)-L-arginyl-[protein] + H2O = lactate + L-arginyl-[protein] + H+
|
|
2.
|
N6-(1-hydroxy-2-oxopropyl)-L-lysyl-[protein] + H2O = lactate + L-lysyl-[protein] + H+
|
|
3.
|
S-(1-hydroxy-2-oxopropyl)-L-cysteinyl-[protein] + H2O = lactate + L-cysteinyl-[protein] + H+
|
|
 |
 |
 |
 |
 |
N(omega)-(1-hydroxy-2-oxopropyl)-L-arginyl-[protein]
|
+
|
H2O
|
=
|
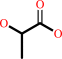 lactate
lactate
|
+
|
L-arginyl-[protein]
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
N(6)-(1-hydroxy-2-oxopropyl)-L-lysyl-[protein]
|
+
|
H2O
|
=
|
 lactate
lactate
|
+
|
L-lysyl-[protein]
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
S-(1-hydroxy-2-oxopropyl)-L-cysteinyl-[protein]
|
+
|
H2O
|
=
|
 lactate
lactate
|
+
|
L-cysteinyl-[protein]
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
ACS Chem Biol
13:2783-2793
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
|
|
S.Tashiro,
J.M.M.Caaveiro,
M.Nakakido,
A.Tanabe,
S.Nagatoishi,
Y.Tamura,
N.Matsuda,
D.Liu,
Q.Q.Hoang,
K.Tsumoto.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
DJ-1 is a Parkinson's disease associated protein endowed with enzymatic, redox
sensing, regulatory, chaperoning, and neuroprotective activities. Although DJ-1
has been vigorously studied for the past decade and a half, its exact role in
the progression of the disease remains uncertain. In addition, little is known
about the spatiotemporal regulation of DJ-1, or the biochemical basis explaining
its numerous biological functions. Progress has been hampered by the lack of
inhibitors with precisely known mechanisms of action. Herein, we have employed
biophysical methodologies and X-ray crystallography to identify and to optimize
a family of compounds inactivating the critical Cys106 residue of human DJ-1. We
demonstrate these compounds are potent inhibitors of various activities of DJ-1
in vitro and in cell-based assays. This study reports a new family of DJ-1
inhibitors with a defined mechanism of action, and contributes toward the
understanding of the biological function of DJ-1.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

