 |
PDBsum entry 6c7e
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of human phosphodiesterase 2a with 1-(2- chlorophenyl)-n,4-dimethyl-[1,2,4]triazolo[4,3-a]quinoxaline-8- carboxamide
|
|
Structure:
|
 |
Cgmp-dependent 3',5'-cyclic phosphodiesterase. Chain: a, b, c, d. Fragment: phosphodiesterase 2a (unp residues 323-661). Synonym: cyclic gmp-stimulated phosphodiesterase, cgspde. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: pde2a. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108.
|
|
Resolution:
|
 |
|
1.43Å
|
R-factor:
|
0.176
|
R-free:
|
0.201
|
|
|
Authors:
|
 |
R.Xu,K.Aertgeerts
|
|
Key ref:
|
 |
L.Gomez
et al.
(2018).
Mathematical and Structural Characterization of Strong Nonadditive Structure-Activity Relationship Caused by Protein Conformational Changes.
J Med Chem,
61,
7754-7766.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
22-Jan-18
|
Release date:
|
15-Aug-18
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O00408
(PDE2A_HUMAN) -
cGMP-dependent 3',5'-cyclic phosphodiesterase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
941 a.a.
342 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.17
- 3',5'-cyclic-nucleotide phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a nucleoside 3',5'-cyclic phosphate + H2O = a nucleoside 5'-phosphate + H+
|
 |
 |
 |
 |
 |
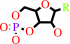 nucleoside 3',5'-cyclic phosphate
nucleoside 3',5'-cyclic phosphate
|
+
|
H2O
|
=
|
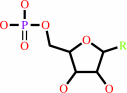 nucleoside 5'-phosphate
nucleoside 5'-phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Med Chem
61:7754-7766
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mathematical and Structural Characterization of Strong Nonadditive Structure-Activity Relationship Caused by Protein Conformational Changes.
|
|
L.Gomez,
R.Xu,
W.Sinko,
B.Selfridge,
W.Vernier,
K.Ly,
R.Truong,
M.Metz,
T.Marrone,
K.Sebring,
Y.Yan,
B.Appleton,
K.Aertgeerts,
M.E.Massari,
J.G.Breitenbucher.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In medicinal chemistry, accurate prediction of additivity-based
structure-activity relationship (SAR) analysis rests on three assumptions: (1) a
consistent binding pose of the central scaffold, (2) no interaction between the
R group substituents, and (3) a relatively rigid binding pocket in which the R
group substituents act independently. Previously, examples of nonadditive SAR
have been documented in systems that deviate from the first two assumptions.
Local protein structural change upon ligand binding, through induced fit or
conformational selection, although a well-known phenomenon that invalidates the
third assumption, has not been linked to nonadditive SAR conclusively. Here, for
the first time, we present clear structural evidence that the formation of a
hydrophobic pocket upon ligand binding in PDE2 catalytic site reduces the size
of another distinct subpocket and contributes to strong nonadditive SAR between
two otherwise distant R groups.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

