 |
PDBsum entry 5y8h

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
5y8h
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.31
- 3-hydroxyisobutyrate dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-hydroxy-2-methylpropanoate + NAD+ = 2-methyl-3-oxopropanoate + NADH + H+
|
 |
 |
 |
 |
 |
3-hydroxy-2-methylpropanoate
Bound ligand (Het Group name = )
matches with 54.55% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
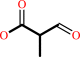 2-methyl-3-oxopropanoate
2-methyl-3-oxopropanoate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochem J
475:2457-2471
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure, interactions and action of Mycobacterium tuberculosis 3-hydroxyisobutyric acid dehydrogenase.
|
|
R.Srikalaivani,
A.Singh,
M.Vijayan,
A.Surolia.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Biochemical and crystallographic studies on Mycobacterium tuberculosis
3-hydroxyisobutyric acid dehydrogenase (MtHIBADH), a member of the
3-hydroxyacid dehydrogenase superfamily, have been carried out. Gel filtration
and blue native PAGE of MtHIBADH show that the enzyme is a dimer. The
enzyme preferentially uses NAD+ as the cofactor and is specific to
S-hydroxyisobutyric acid (HIBA). It can also use R-HIBA, l-serine
and 3-hydroxypropanoic acid (3-HP) as substrates, but with much less efficiency.
The pH optimum for activity is ∼11. Structures of the native enzyme, the
holoenzyme, binary complexes with NAD+, S-HIBA, R-HIBA,
l-serine and 3-HP and ternary complexes involving the substrates and
NAD+ have been determined. None of the already known structures of
HIBADH contain a substrate molecule at the binding site. The structures reported
here provide for the first time, among other things, a clear indication of the
location and interactions of the substrates at the active site. They also define
the entrance of the substrates to the active site region. The structures provide
information on the role of specific residues at the active site and the
entrance. The results obtained from crystal structures are consistent with
solution studies including mutational analysis. They lead to the proposal of a
plausible mechanism of the action of the enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

